Crystal structure of ribose-5-phosphate isomerase A from Ochrobactrum sp. CSL1
Xu, X.Q., Mo, X.B., Ju, X., Shen, M., Li, L.Z., Yuan, A.Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribose-5-phosphate isomerase A | 232 | Ochrobactrum sp. | Mutation(s): 0 EC: 5.3.1.6 | 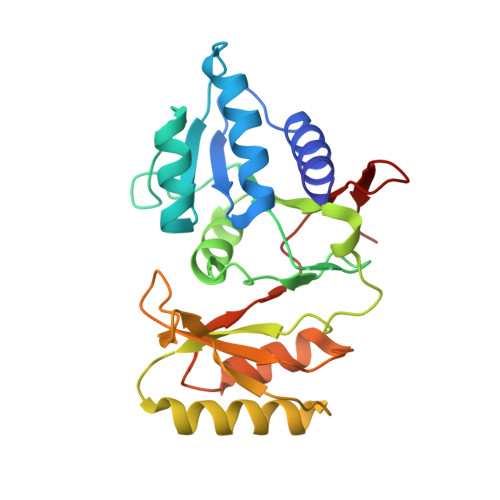 | |
UniProt | |||||
Find proteins for A6X0Q4 (Brucella anthropi (strain ATCC 49188 / DSM 6882 / CCUG 24695 / JCM 21032 / LMG 3331 / NBRC 15819 / NCTC 12168 / Alc 37)) Explore A6X0Q4 Go to UniProtKB: A6X0Q4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A6X0Q4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | C [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 69.817 | α = 90 |
| b = 169.262 | β = 90 |
| c = 41.268 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data collection |
| HKL-2000 | data scaling |
| HKL-2000 | data reduction |
| SHARP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China | China | 21376156 |
| National Natural Science Foundation of China | China | 21676173 |