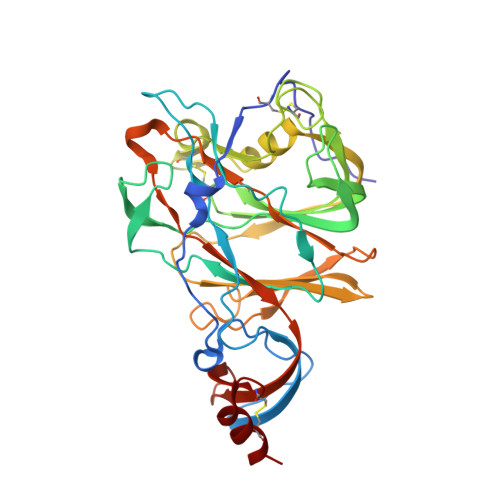
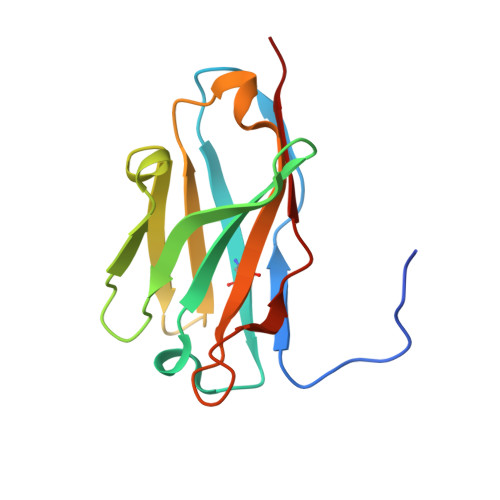
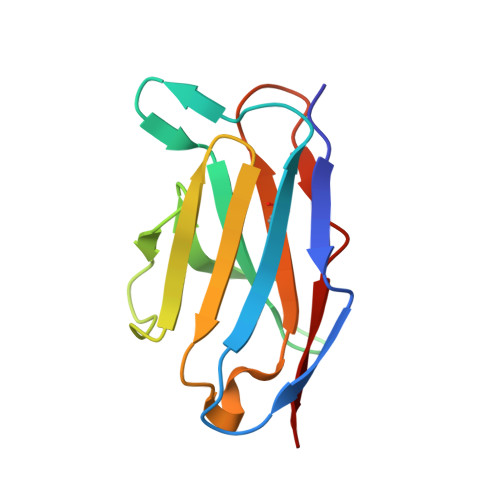
Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein.
Zhou, H., Chen, Y., Zhang, S., Niu, P., Qin, K., Jia, W., Huang, B., Zhang, S., Lan, J., Zhang, L., Tan, W., Wang, X.(2019) Nat Commun 10: 3068-3068
- PubMed: 31296843
- DOI: https://doi.org/10.1038/s41467-019-10897-4
- Primary Citation of Related Structures:
6J11 - PubMed Abstract:
Most neutralizing antibodies against Middle East respiratory syndrome coronavirus (MERS-CoV) target the receptor-binding domain (RBD) of the spike glycoprotein and block its binding to the cellular receptor dipeptidyl peptidase 4 (DPP4). The epitopes and mechanisms of mAbs targeting non-RBD regions have not been well characterized yet. Here we report the monoclonal antibody 7D10 that binds to the N-terminal domain (NTD) of the spike glycoprotein and inhibits the cell entry of MERS-CoV with high potency. Structure determination and mutagenesis experiments reveal the epitope and critical residues on the NTD for 7D10 binding and neutralization. Further experiments indicate that the neutralization by 7D10 is not solely dependent on the inhibition of DPP4 binding, but also acts after viral cell attachment, inhibiting the pre-fusion to post-fusion conformational change of the spike. These properties give 7D10 a wide neutralization breadth and help explain its synergistic effects with several RBD-targeting antibodies.
- The Ministry of Education Key Laboratory of Protein Science, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Collaborative Innovation Center for Biotherapy, School of Life Sciences, Tsinghua University, 100084, Beijing, China.
Organizational Affiliation: