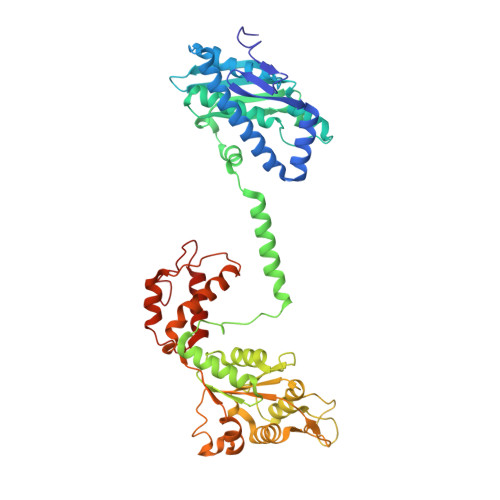
Tetrameric architecture of an active phenol-bound form of the AAA+transcriptional regulator DmpR.
Park, K.H., Kim, S., Lee, S.J., Cho, J.E., Patil, V.V., Dumbrepatil, A.B., Song, H.N., Ahn, W.C., Joo, C., Lee, S.G., Shingler, V., Woo, E.J.(2020) Nat Commun 11: 2728-2728
- PubMed: 32483114
- DOI: https://doi.org/10.1038/s41467-020-16562-5
- Primary Citation of Related Structures:
6IY8 - PubMed Abstract:
The Pseudomonas putida phenol-responsive regulator DmpR is a bacterial enhancer binding protein (bEBP) from the AAA + ATPase family. Even though it was discovered more than two decades ago and has been widely used for aromatic hydrocarbon sensing, the activation mechanism of DmpR has remained elusive. Here, we show that phenol-bound DmpR forms a tetramer composed of two head-to-head dimers in a head-to-tail arrangement. The DmpR-phenol complex exhibits altered conformations within the C-termini of the sensory domains and shows an asymmetric orientation and angle in its coiled-coil linkers. The structural changes within the phenol binding sites and the downstream ATPase domains suggest that the effector binding signal is propagated through the coiled-coil helixes. The tetrameric DmpR-phenol complex interacts with the σ 54 subunit of RNA polymerase in presence of an ATP analogue, indicating that DmpR-like bEBPs tetramers utilize a mechanistic mode distinct from that of hexameric AAA + ATPases to activate σ 54 -dependent transcription.
- Disease Target Structure Research Center, Korea Research Institute of Bioscience & Biotechnology (KRIBB), Daejeon, 305-806, Republic of Korea.
Organizational Affiliation: