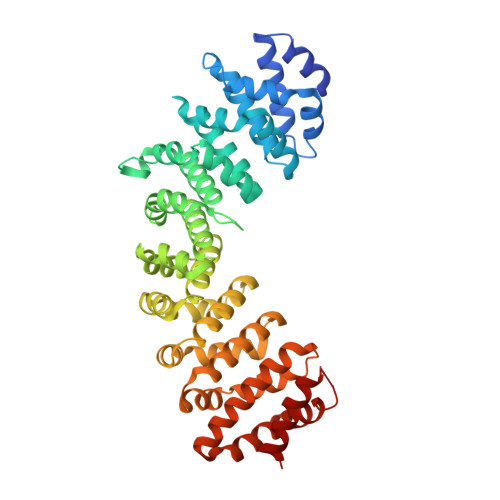
Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-alpha.
Chang, C.-C., Chen, C.-J., Grauffel, C., Pien, Y.-C., Lim, C., Tsai, S.-Y., Hsia, K.-C.(2019) Nat Commun 10: 4307-4307
- PubMed: 31541088
- DOI: https://doi.org/10.1038/s41467-019-12207-4
- Primary Citation of Related Structures:
6IW8, 6IWA, 6K06 - PubMed Abstract:
To facilitate proper mitotic cell partitioning, the Golgi disassembles by suppressing vesicle fusion. However, the underlying mechanism has not been characterized previously. Here, we report a Ran pathway-independent attenuation mechanism that allows Importin-α (a nuclear transport factor) to suppress the vesicle fusion mediated by p115 (a vesicular tethering factor) and is required for mitotic Golgi disassembly. We demonstrate that Importin-α directly competes with p115 for interaction with the Golgi protein GM130. This interaction, promoted by a phosphate moiety on GM130, is independent of Importin-β and Ran. A GM130 K34A mutant, in which the Importin-α-GM130 interaction is specifically disrupted, exhibited abundant Golgi puncta during metaphase. Importantly, a mutant showing enhanced p115-GM130 interaction presented proliferative defects and G2/M arrest, demonstrating that Importin-α-GM130 binding modulates the Golgi disassembly that governs mitotic progression. Our findings illuminate that the Ran and kinase-phosphatase pathways regulate multiple aspects of mitosis coordinated by Importin-α (e.g. spindle assembly, Golgi disassembly).
- Institute of Molecular Biology, Academia Sinica, Taipei, 11529, Taiwan.
Organizational Affiliation: