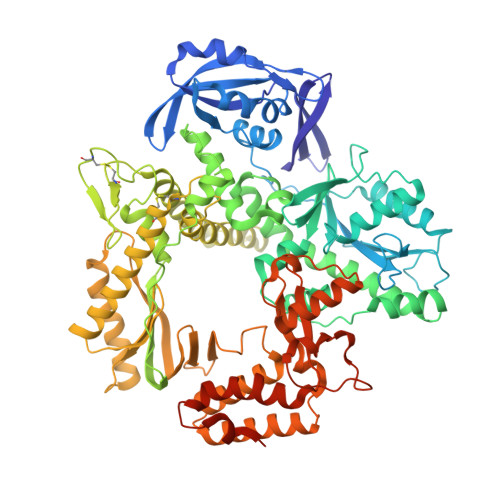
Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
LinWu, S.W., Tu, Y.H., Tsai, T.Y., Maestre-Reyna, M., Liu, M.S., Wu, W.J., Huang, J.Y., Chi, H.W., Chang, W.H., Chiou, C.F., Wang, A.H., Lee, J., Tsai, M.D.(2019) Commun Biol 2: 224-224
- PubMed: 31240262
- DOI: https://doi.org/10.1038/s42003-019-0458-7
- Primary Citation of Related Structures:
6IS7, 6ISF, 6ISG, 6ISH, 6ISI - PubMed Abstract:
It was reported in 1995 that T7 and Taq DNA polymerases possess 3'-esterase activity, but without follow-up studies. Here we report that the 3'-esterase activity is intrinsic to the Thermococcus sp . 9°N DNA polymerase, and that it can be developed into a continuous method for DNA sequencing with dNTP analogs carrying a 3'-ester with a fluorophore. We first show that 3'-esterified dNTP can be incorporated into a template-primer DNA, and solve the crystal structures of the reaction intermediates and products. Then we show that the reaction can occur continuously, modulated by active site residues Tyr409 and Asp542. Finally, we use 5'-FAM-labeled primer and esterified dNTP with a dye to show that the reaction can proceed to ca. 450 base pairs, and that the intermediates of many individual steps can be identified. The results demonstrate the feasibility of a 3'-editing based DNA sequencing method that could find practical applications after further optimization.
- Personal Genomics, Inc., Zhubei, Hsinchu 30261 Taiwan.
Organizational Affiliation: