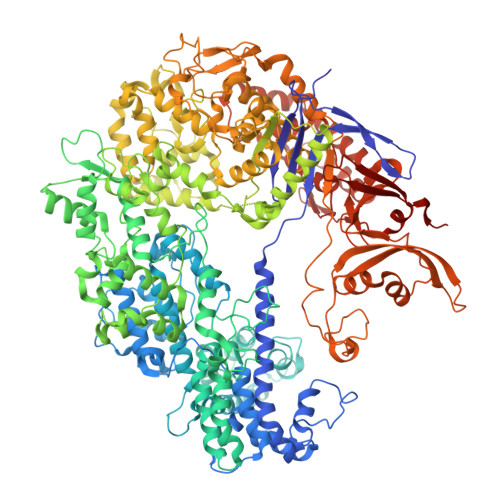
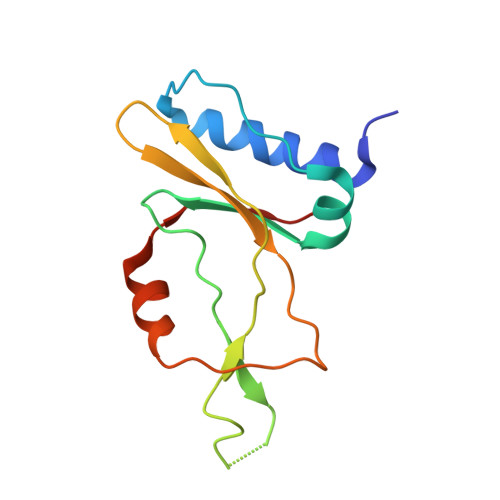
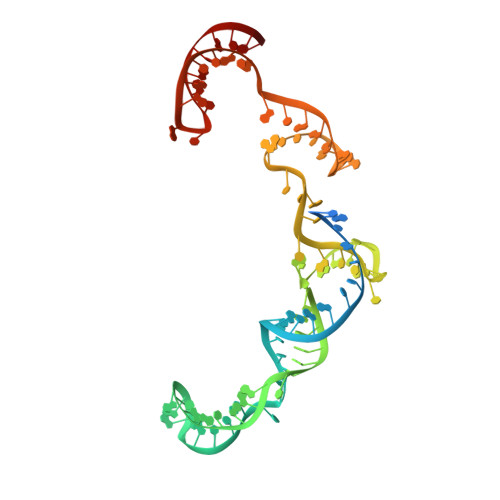
Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race.
Liu, L., Yin, M., Wang, M., Wang, Y.(2019) Mol Cell 73: 611-620.e3
- PubMed: 30606466
- DOI: https://doi.org/10.1016/j.molcel.2018.11.011
- Primary Citation of Related Structures:
6IFO - PubMed Abstract:
CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated proteins) systems provide prokaryotic cells with adaptive immunity against invading bacteriophages. Bacteriophages counteract bacterial responses by encoding anti-CRISPR inhibitor proteins (Acr). However, the structural basis for their inhibitory actions remains largely unknown. Here, we report the crystal structure of the AcrIIA2-SpyCas9-sgRNA (single-guide RNA) complex at 3.3 Å resolution. We show that AcrIIA2 binds SpyCas9 at a position similar to the target DNA binding region. More specifically, AcrIIA2 interacts with the protospacer adjacent motif (PAM) recognition residues of Cas9, preventing target double-stranded DNA (dsDNA) detection. Thus, phage-encoded AcrIIA2 appears to act as a DNA mimic that blocks subsequent dsDNA binding by virtue of its highly acidic residues, disabling bacterial Cas9 by competing with target dsDNA binding with a binding motif distinct from AcrIIA4. Our study provides a more detailed mechanistic understanding of AcrIIA2-mediated inhibition of SpyCas9, the most widely used genome-editing tool, opening new avenues for improved regulatory precision during genome editing.
- Key Laboratory of RNA Biology, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: