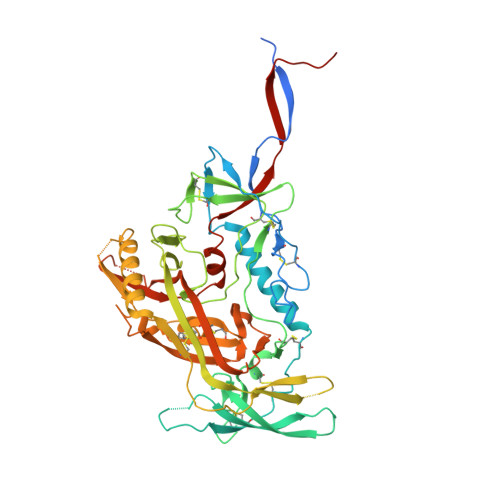
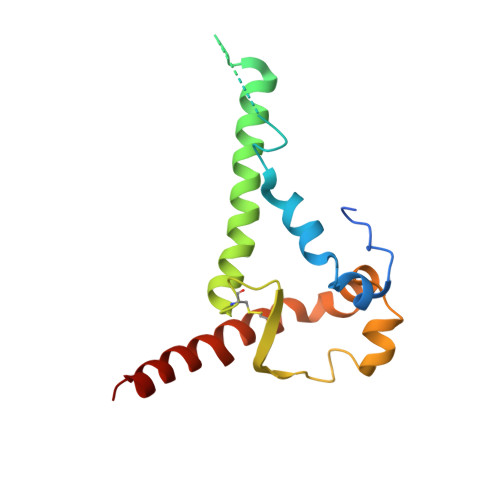
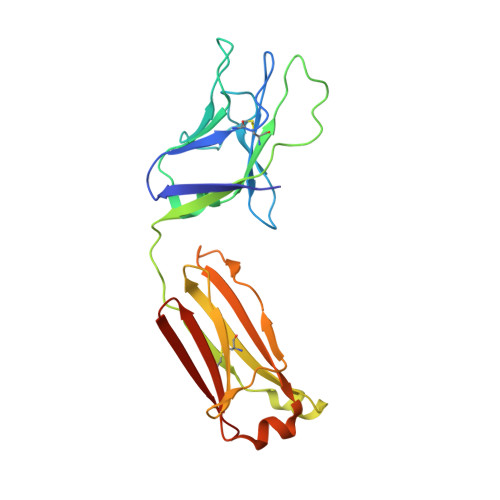
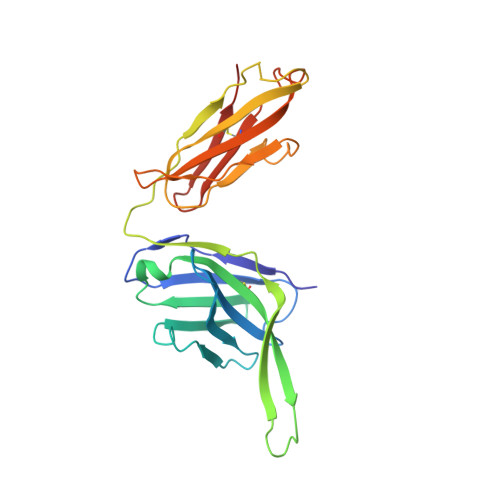
Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence.
Sliepen, K., Han, B.W., Bontjer, I., Mooij, P., Garces, F., Behrens, A.J., Rantalainen, K., Kumar, S., Sarkar, A., Brouwer, P.J.M., Hua, Y., Tolazzi, M., Schermer, E., Torres, J.L., Ozorowski, G., van der Woude, P., de la Pena, A.T., van Breemen, M.J., Camacho-Sanchez, J.M., Burger, J.A., Medina-Ramirez, M., Gonzalez, N., Alcami, J., LaBranche, C., Scarlatti, G., van Gils, M.J., Crispin, M., Montefiori, D.C., Ward, A.B., Koopman, G., Moore, J.P., Shattock, R.J., Bogers, W.M., Wilson, I.A., Sanders, R.W.(2019) Nat Commun 10: 2355-2355
- PubMed: 31142746
- DOI: https://doi.org/10.1038/s41467-019-10262-5
- Primary Citation of Related Structures:
6IEQ - PubMed Abstract:
Stabilized HIV-1 envelope glycoproteins (Env) that resemble the native Env are utilized in vaccination strategies aimed at inducing broadly neutralizing antibodies (bNAbs). To limit the exposure of rare isolate-specific antigenic residues/determinants we generated a SOSIP trimer based on a consensus sequence of all HIV-1 group M isolates (ConM). The ConM trimer displays the epitopes of most known bNAbs and several germline bNAb precursors. The crystal structure of the ConM trimer at 3.9 Å resolution resembles that of the native Env trimer and its antigenic surface displays few rare residues. The ConM trimer elicits strong NAb responses against the autologous virus in rabbits and macaques that are significantly enhanced when it is presented on ferritin nanoparticles. The dominant NAb specificity is directed against an epitope at or close to the trimer apex. Immunogens based on consensus sequences might have utility in engineering vaccines against HIV-1 and other viruses.
- Department of Medical Microbiology, Amsterdam Infection & Immunity Institute, Amsterdam UMC, University of Amsterdam, Meibergdreef 9, Amsterdam, 1105AZ, The Netherlands.
Organizational Affiliation: