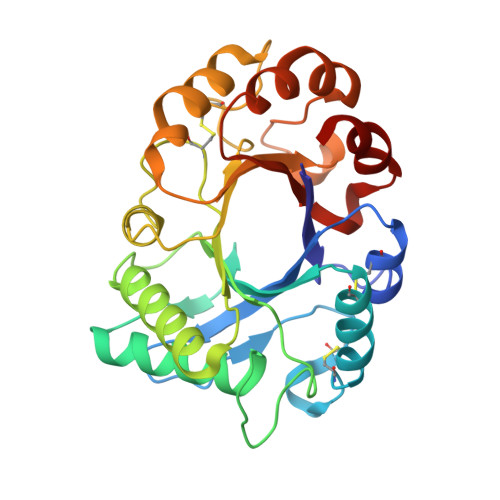
TIM barrel fold and glycan moieties in the structure of ICChI, a protein with chitinase and lysozyme activity.
Kumar, S., Kumar, A., Patel, A.K.(2020) Phytochemistry 170: 112221-112221
- PubMed: 31790908
- DOI: https://doi.org/10.1016/j.phytochem.2019.112221
- Primary Citation of Related Structures:
6IDN - PubMed Abstract:
The ICChI is a 35-kDa, glycosylated protein isolated from the latex of the weed Ipomoea carnea. It displays chitinase and lysozyme activity, which could be important for the defense against pathogenic fungi, insects and bacteria. The ICChI enzyme was crystallized, and a diffraction data set was collected from a single crystal to 1.42 Å resolution. The crystals belong to the primitive tetragonal space group P4 3 2 1 2, with unit-cell parameters a = b = 57.9, c = 172.0 Å, and α = β = γ = 90°. The structure was elucidated by molecular replacement method using a mixed model of three homologous structures from the N-terminal sequence of ICChI. The refined model consists of 272 amino acid residues and has a R factor of 18.93% and R free of 22.42%. The protein consists of a single globular domain with a (α/β) 8 triosephosphate isomerase barrel fold. Three of the consensus sites for N-glycosylation viz., Asn 45 , Asn 172 , and Asn 194 containing carbohydrate moieties N-Acetylglucosamine (NAG), mannose, fucose, and xylose. The putative catalytic residues are Asp 125 , Glu 127 , and Tyr 184 . The crystal structure may provide fundamental information of GH18 family chitinases.
- Kusuma School of Biological Sciences, Indian Institute of Technology Delhi, New Delhi, 110016, India.
Organizational Affiliation: