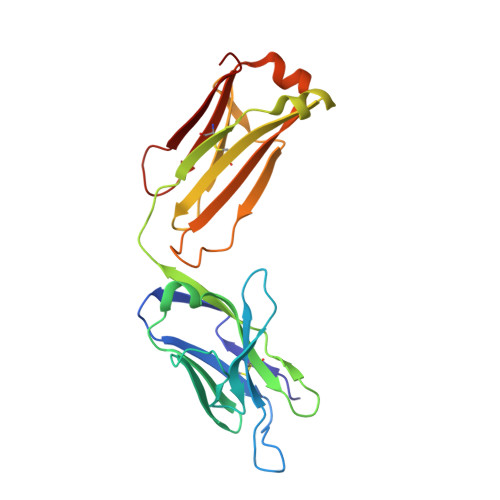
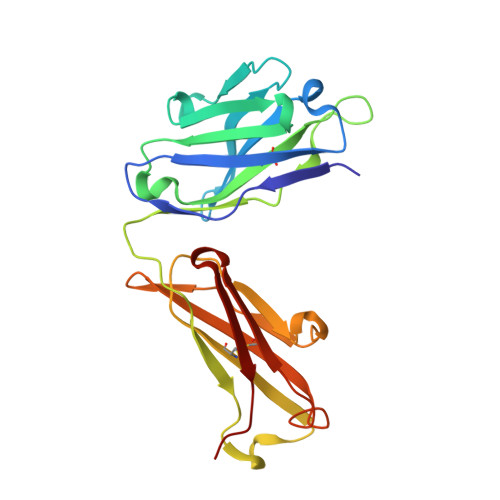
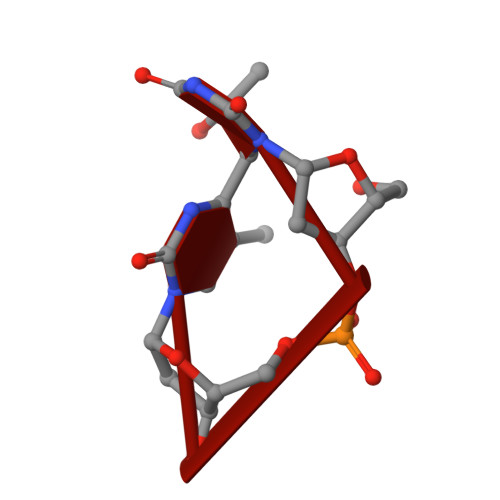
Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Yokoyama, H., Mizutani, R., Noguchi, S., Hayashida, N.(2019) Acta Crystallogr F Struct Biol Commun 75: 80-88
- PubMed: 30713158
- DOI: https://doi.org/10.1107/S2053230X18017661
- Primary Citation of Related Structures:
6IDG, 6IDH - PubMed Abstract:
DNA photoproducts with (6-4) pyrimidine-pyrimidone adducts produced by ultraviolet light are mutagenic and carcinogenic. The crystal structures of the anti-(6-4) photoproduct antibody 64M-5 Fab and of its complex with dT(6-4)T were determined at 2.5 and 2.0 Å resolution, respectively. A comparison between the dT(6-4)T-liganded and unliganded structures indicates that the side chain of His93L is greatly rotated and shifted on binding to dT(6-4)T, leading to the formation of an electrostatic interaction with the phosphate moiety of dT(6-4)T, which shows a remarkable induced fit. Based on a comparison of the dT(6-4)T-liganded structures of the 64M-5 and 64M-2 Fabs, the electrostatic interaction between the side chain of His93L in 64M-5 and the phosphate moiety of dT(6-4)T is lost for Leu93L in 64M-2, while Arg90L in 64M-5 instead of Gln90L in 64M-2 stabilizes the conformation of complementarity-determining region (CDR) L3. These differences contribute to the higher affinity of 64M-5 for dT(6-4)T compared with that of 64M-2.
- Faculty of Pharmaceutical Sciences, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278-8510, Japan.
Organizational Affiliation: