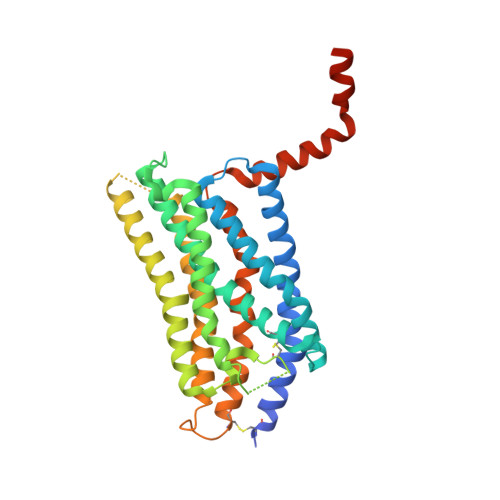
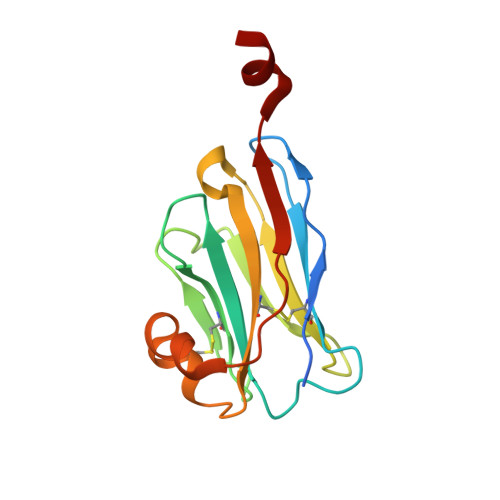
Structural basis of species-selective antagonist binding to the succinate receptor.
Haffke, M., Fehlmann, D., Rummel, G., Boivineau, J., Duckely, M., Gommermann, N., Cotesta, S., Sirockin, F., Freuler, F., Littlewood-Evans, A., Kaupmann, K., Jaakola, V.P.(2019) Nature 574: 581-585
- PubMed: 31645725
- DOI: https://doi.org/10.1038/s41586-019-1663-8
- Primary Citation of Related Structures:
6IBB, 6RNK - PubMed Abstract:
The tricarboxylic acid cycle intermediate succinate is involved in metabolic processes and plays a crucial role in the homeostasis of mitochondrial reactive oxygen species 1 . The receptor responsible for succinate signalling, SUCNR1 (also known as GPR91), is a member of the G-protein-coupled-receptor family 2 and links succinate signalling to renin-induced hypertension, retinal angiogenesis and inflammation 3-5 . Because SUCNR1 senses succinate as an immunological danger signal 6 -which has relevance for diseases including ulcerative colitis, liver fibrosis 7 , diabetes and rheumatoid arthritis 3,8 -it is of interest as a therapeutic target. Here we report the high-resolution crystal structure of rat SUCNR1 in complex with an intracellular binding nanobody in the inactive conformation. Structure-based mutagenesis and radioligand-binding studies, in conjunction with molecular modelling, identified key residues for species-selective antagonist binding and enabled the determination of the high-resolution crystal structure of a humanized rat SUCNR1 in complex with a high-affinity, human-selective antagonist denoted NF-56-EJ40. We anticipate that these structural insights into the architecture of the succinate receptor and its antagonist selectivity will enable structure-based drug discovery and will further help to elucidate the function of SUCNR1 in vitro and in vivo.
- Chemical Biology & Therapeutics, Novartis Institutes for BioMedical Research, Novartis Pharma AG, Basel, Switzerland. matthias.haffke@novartis.com.
Organizational Affiliation: