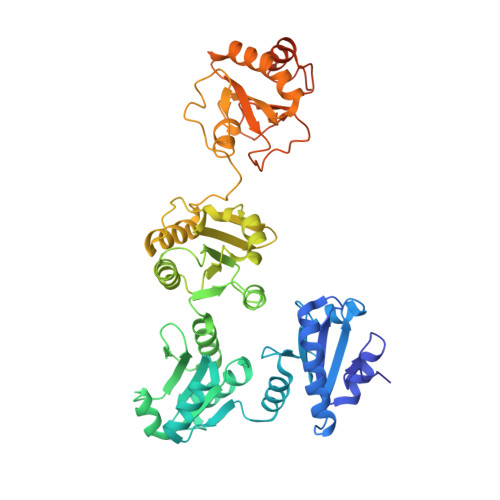
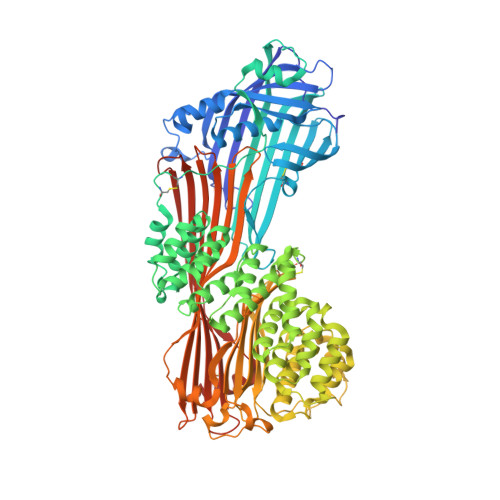
The crystal structure of human microsomal triglyceride transfer protein.
Biterova, E.I., Isupov, M.N., Keegan, R.M., Lebedev, A.A., Sohail, A.A., Liaqat, I., Alanen, H.I., Ruddock, L.W.(2019) Proc Natl Acad Sci U S A 116: 17251-17260
- PubMed: 31395737
- DOI: https://doi.org/10.1073/pnas.1903029116
- Primary Citation of Related Structures:
6I7S - PubMed Abstract:
Microsomal triglyceride transfer protein (MTP) plays an essential role in lipid metabolism, especially in the biogenesis of very low-density lipoproteins and chylomicrons via the transfer of neutral lipids and the assembly of apoB-containing lipoproteins. Our understanding of the molecular mechanisms of MTP has been hindered by a lack of structural information of this heterodimeric complex comprising an MTPα subunit and a protein disulfide isomerase (PDI) β-subunit. The structure of MTP presented here gives important insights into the potential mechanisms of action of this essential lipid transfer molecule, structure-based rationale for previously reported disease-causing mutations, and a means for rational drug design against cardiovascular disease and obesity. In contrast to the previously reported structure of lipovitellin, which has a funnel-like lipid-binding cavity, the lipid-binding site is encompassed in a β-sandwich formed by 2 β-sheets from the C-terminal domain of MTPα. The lipid-binding cavity of MTPα is large enough to accommodate a single lipid. PDI independently has a major role in oxidative protein folding in the endoplasmic reticulum. Comparison of the mechanism of MTPα binding by PDI with previously published structures gives insights into large protein substrate binding by PDI and suggests that the previous structures of human PDI represent the "substrate-bound" and "free" states rather than differences arising from redox state.
- Faculty of Biochemistry and Molecular Medicine, University of Oulu, 90220 Oulu, Finland.
Organizational Affiliation: