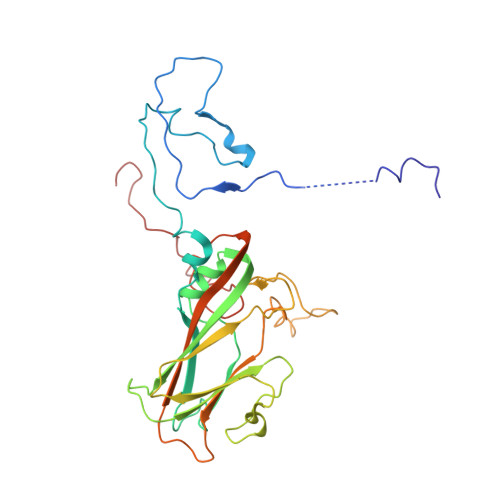
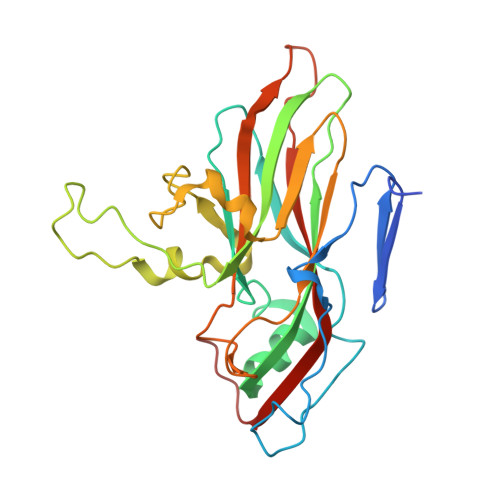
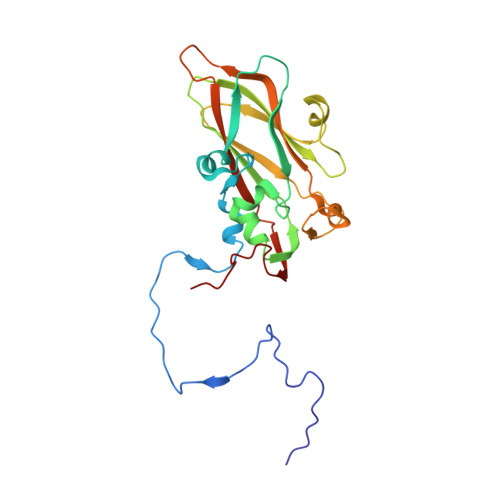
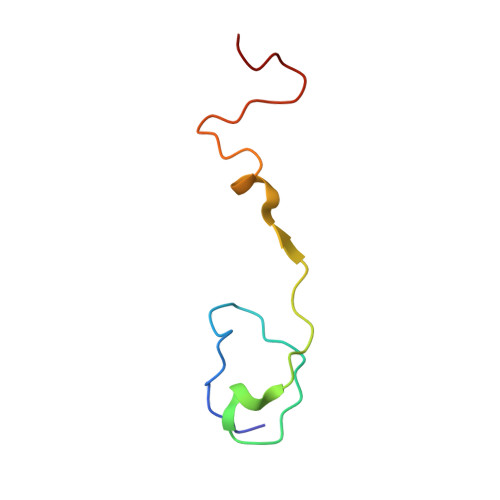
Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Zhou, D., Zhao, Y., Kotecha, A., Fry, E.E., Kelly, J.T., Wang, X., Rao, Z., Rowlands, D.J., Ren, J., Stuart, D.I.(2019) Nat Microbiol 4: 414-419
- PubMed: 30531980
- DOI: https://doi.org/10.1038/s41564-018-0319-z
- Primary Citation of Related Structures:
6I2K - PubMed Abstract:
Enterovirus 71 (EV71) is a common cause of hand, foot and mouth disease-a disease endemic especially in the Asia-Pacific region 1 . Scavenger receptor class B member 2 (SCARB2) is the major receptor of EV71, as well as several other enteroviruses responsible for hand, foot and mouth disease, and plays a key role in cell entry 2 . The isolated structures of EV71 and SCARB2 are known 3-6 , but how they interact to initiate infection is not. Here, we report the EV71-SCARB2 complex structure determined at 3.4 Å resolution using cryo-electron microscopy. This reveals that SCARB2 binds EV71 on the southern rim of the canyon, rather than across the canyon, as predicted 3,7,8 . Helices 152-163 (α5) and 183-193 (α7) of SCARB2 and the viral protein 1 (VP1) GH and VP2 EF loops of EV71 dominate the interaction, suggesting an allosteric mechanism by which receptor binding might facilitate the low-pH uncoating of the virus in the endosome/lysosome. Remarkably, many residues within the binding footprint are not conserved across SCARB2-dependent enteroviruses; however, a conserved proline and glycine seem to be key residues. Thus, although the virus maintains antigenic variability even within the receptor-binding footprint, the identification of binding 'hot spots' may facilitate the design of receptor mimic therapeutics less likely to quickly generate resistance.
- Division of Structural Biology, The Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK.
Organizational Affiliation: