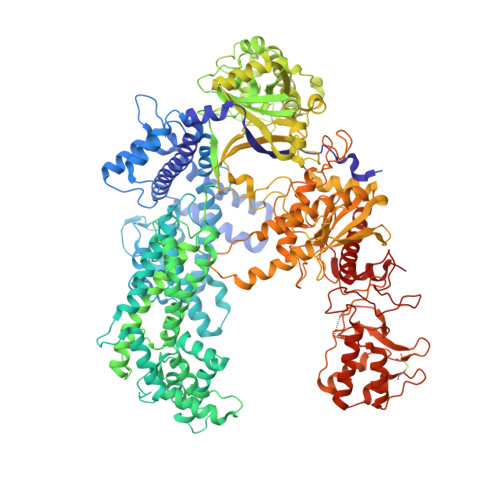
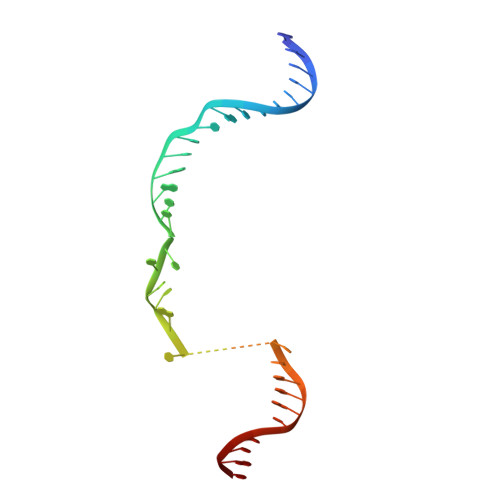
Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a.
Swarts, D.C., Jinek, M.(2019) Mol Cell 73: 589-600.e4
- PubMed: 30639240
- DOI: https://doi.org/10.1016/j.molcel.2018.11.021
- Primary Citation of Related Structures:
6I1K, 6I1L - PubMed Abstract:
CRISPR-Cas12a (Cpf1) is an RNA-guided DNA-cutting nuclease that has been repurposed for genome editing. Upon target DNA binding, Cas12a cleaves both the target DNA in cis and non-target single-stranded DNAs (ssDNAs) in trans. To elucidate the molecular basis for both DNase cleavage modes, we performed structural and biochemical studies on Francisella novicida Cas12a. We show that guide RNA-target strand DNA hybridization conformationally activates Cas12a, triggering its trans-acting, non-specific, single-stranded DNase activity. In turn, cis cleavage of double-stranded DNA targets is a result of protospacer adjacent motif (PAM)-dependent DNA duplex unwinding, electrostatic stabilization of the displaced non-target DNA strand, and ordered sequential cleavage of the non-target and target DNA strands. Cas12a releases the PAM-distal DNA cleavage product and remains bound to the PAM-proximal DNA cleavage product in a catalytically competent, trans-active state. Together, these results provide a revised model for the molecular mechanisms of both the cis- and the trans-acting DNase activities of Cas12a enzymes, enabling their further exploitation as genome editing tools.
- Department of Biochemistry, University of Zurich, 8057 Zurich, Switzerland.
Organizational Affiliation: