Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Felisberto-Rodrigues, C., Thomas, J.C., McAndrew, C., Le Bihan, Y.V., Burke, R., Workman, P., van Montfort, R.L.M.(2019) Biochem J 476: 2521-2543
- PubMed: 31409651
- DOI: https://doi.org/10.1042/BCJ20190383
- Primary Citation of Related Structures:
6HYS, 6HYT, 6HYU - PubMed Abstract:
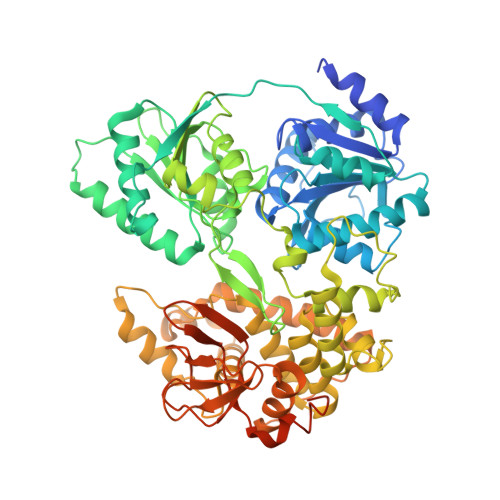
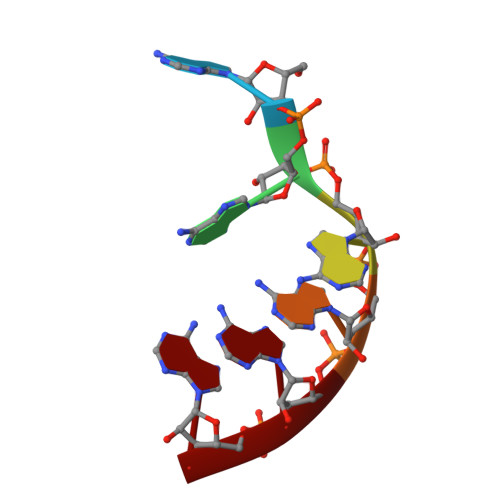
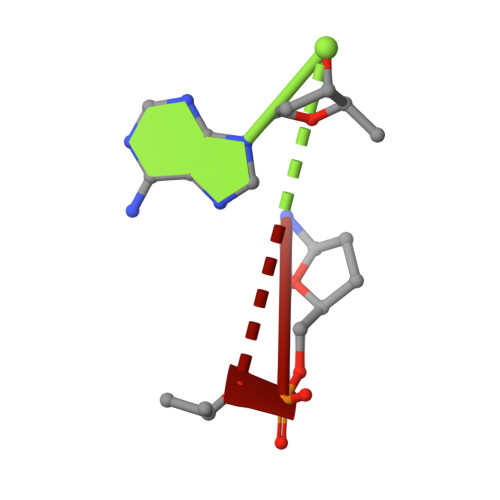
DHX8 is a crucial DEAH-box RNA helicase involved in splicing and required for the release of mature mRNA from the spliceosome. Here, we report the biochemical characterisation of full-length human DHX8 and the catalytically active helicase core DHX8Δ547, alongside crystal structures of DHX8Δ547 bound to ADP and a structure of DHX8Δ547 bound to poly(A) 6 single-strand RNA. Our results reveal that DHX8 has an in vitro binding preference for adenine-rich RNA and that RNA binding triggers the release of ADP through significant conformational flexibility in the conserved DEAH-, P-loop and hook-turn motifs. We demonstrate the importance of R620 and both the hook-turn and hook-loop regions for DHX8 helicase activity and propose that the hook-turn acts as a gatekeeper to regulate the directional movement of the 3' end of RNA through the RNA-binding channel. This study provides an in-depth understanding of the activity of DHX8 and contributes insights into the RNA-unwinding mechanisms of the DEAH-box helicase family.
- Cancer Research UK Cancer Therapeutics Unit, Division of Cancer Therapeutics, The Institute of Cancer Research, London SM2 5NG, U.K.
Organizational Affiliation: