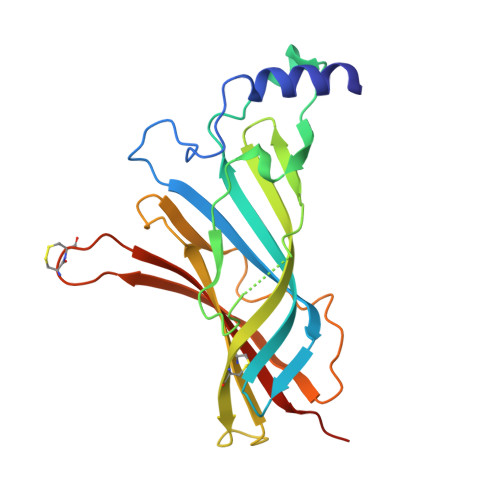
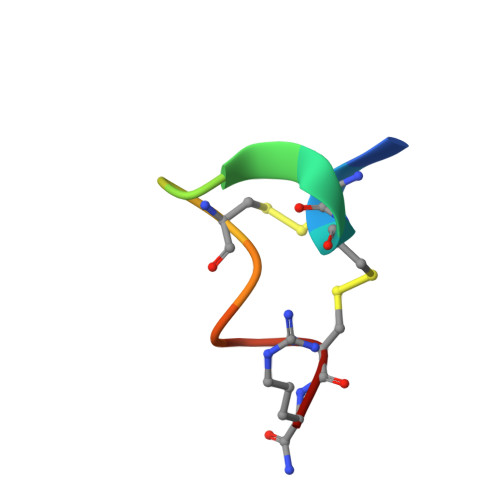
Crystal Structure of the Monomeric Extracellular Domain of alpha 9 Nicotinic Receptor Subunit in Complex With alpha-Conotoxin RgIA: Molecular Dynamics Insights Into RgIA Binding to alpha 9 alpha 10 Nicotinic Receptors.
Zouridakis, M., Papakyriakou, A., Ivanov, I.A., Kasheverov, I.E., Tsetlin, V., Tzartos, S., Giastas, P.(2019) Front Pharmacol 10: 474-474
- PubMed: 31118896
- DOI: https://doi.org/10.3389/fphar.2019.00474
- Primary Citation of Related Structures:
6HY7 - PubMed Abstract:
The α9 subunit of nicotinic acetylcholine receptors (nAChRs) exists mainly in heteropentameric assemblies with α10. Accumulating data indicate the presence of three different binding sites in α9α10 nAChRs: the α9(+)/α9(-), the α9(+)/α10(-), and the α10(+)/α9(-). The major role of the principal (+) side of the extracellular domain (ECD) of α9 subunit in binding of the antagonists methyllylcaconitine and α-bungarotoxin was shown previously by the crystal structures of the monomeric α9-ECD with these molecules. Here we present the 2.26-Å resolution crystal structure of α9-ECD in complex with α-conotoxin (α-Ctx) RgIA, a potential drug for chronic pain, the first structure reported for a complex between an nAChR domain and an α-Ctx. Superposition of this structure with those of other α-Ctxs bound to the homologous pentameric acetylcholine binding proteins revealed significant similarities in the orientation of bound conotoxins, despite the monomeric state of the α9-ECD. In addition, ligand-binding studies calculated a binding affinity of RgIA to the α9-ECD at the low micromolar range. Given the high identity between α9 and α10 ECDs, particularly at their (+) sides, the presented structure was used as template for molecular dynamics simulations of the ECDs of the human α9α10 nAChR in pentameric assemblies. Our results support a favorable binding of RgIA at α9(+)/α9(-) or α10(+)/α9(-) rather than the α9(+)/α10(-) interface, in accordance with previous mutational and functional data.
- Department of Neurobiology, Hellenic Pasteur Institute, Athens, Greece.
Organizational Affiliation: