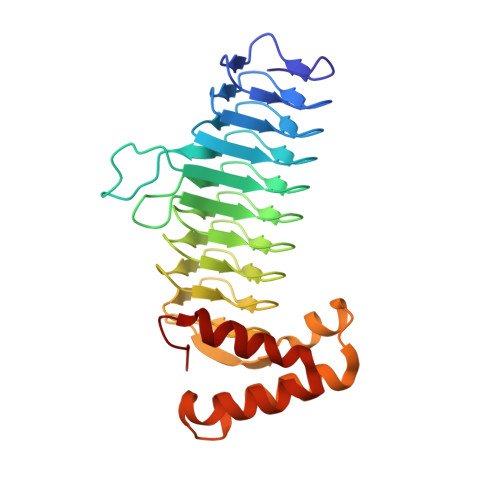
Structure guided design of an antibacterial peptide that targets UDP-N-acetylglucosamine acyltransferase.
Dangkulwanich, M., Raetz, C.R.H., Williams, A.H.(2019) Sci Rep 9: 3947-3947
- PubMed: 30850651
- DOI: https://doi.org/10.1038/s41598-019-40418-8
- Primary Citation of Related Structures:
6HY2 - PubMed Abstract:
UDP-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (LpxA) catalyzes the first step of lipid A biosynthesis, the transfer of an R-3-hydroxyacyl chain from its acyl carrier protein (ACP) to the 3-OH group of UDP-GlcNAc. Essential in the growth of Gram-negative bacteria, LpxA is a logical target for antibiotics design. A pentadecapeptide (Peptide 920) with high affinity towards LpxA was previously identified in a phage display library. Here we created a small library of systematically designed peptides with the length of four to thirteen amino acids using Peptide 920 as a scaffold. The concentrations of these peptides at which 50% of LpxA is inhibited (IC 50 ) range from 50 nM to >100 μM. We determined the crystal structure of E. coli LpxA in a complex with a potent inhibitor. LpxA-inhibitor interaction, solvent model and all contributing factors to inhibitor efficacy were well resolved. The peptide primarily occludes the ACP binding site of LpxA. Interactions between LpxA and the inhibitor are different from those in the structure of Peptide 920. The inhibitory peptide library and the crystal structure of inhibitor-bound LpxA described here may further assist in the rational design of inhibitors with antimicrobial activity that target LpxA and potentially other acyltransferases.
- Department of Biochemistry, Duke University Medical Center, Box 3711, Durham, North Carolina, 27710, USA.
Organizational Affiliation: