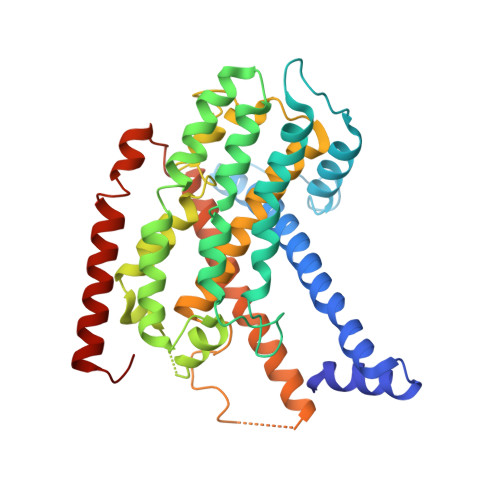
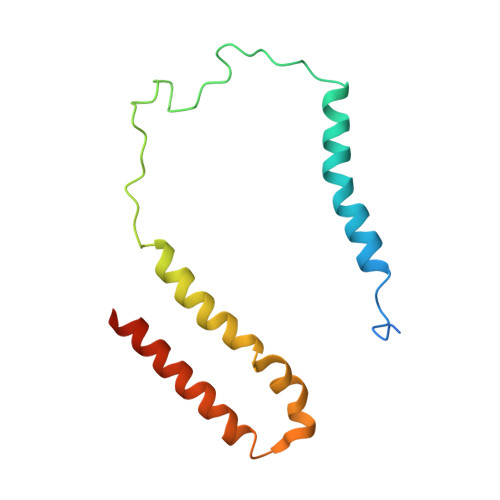
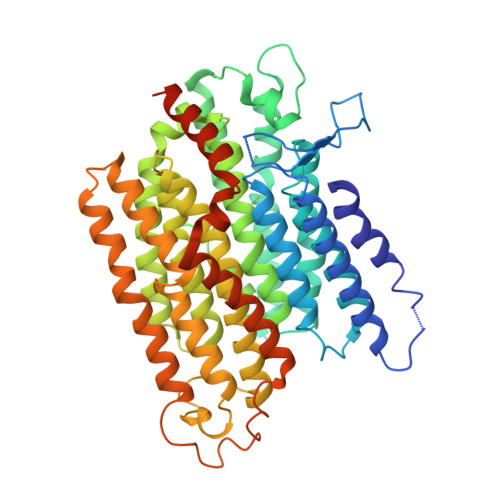
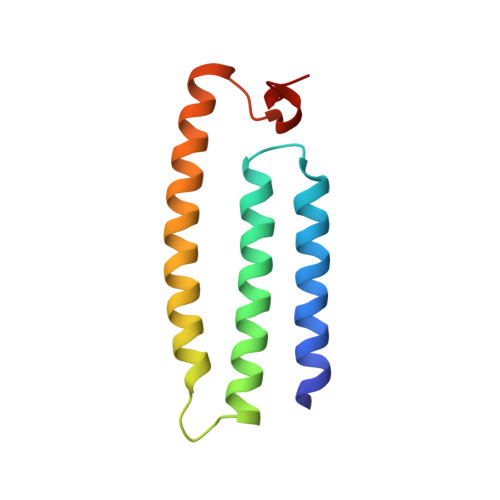
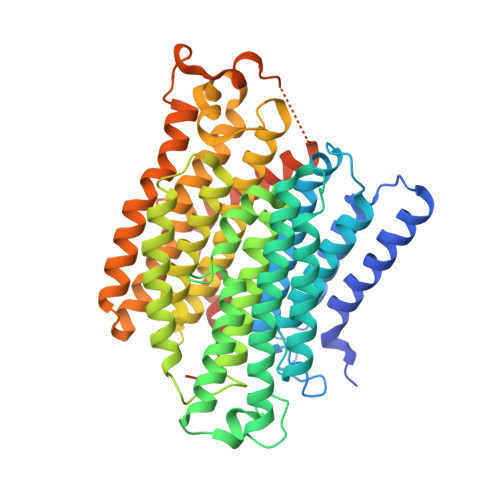
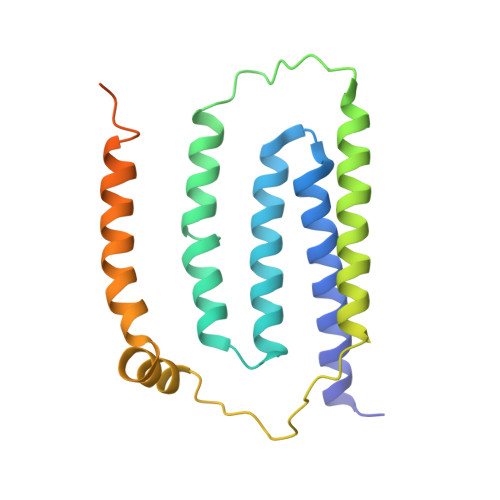
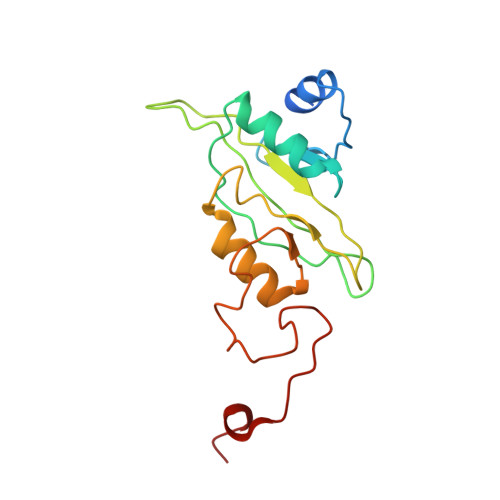
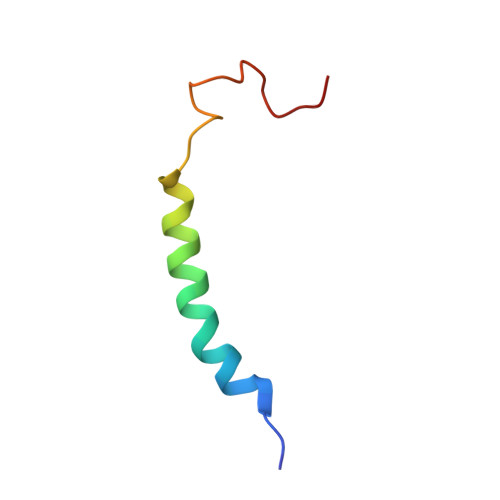
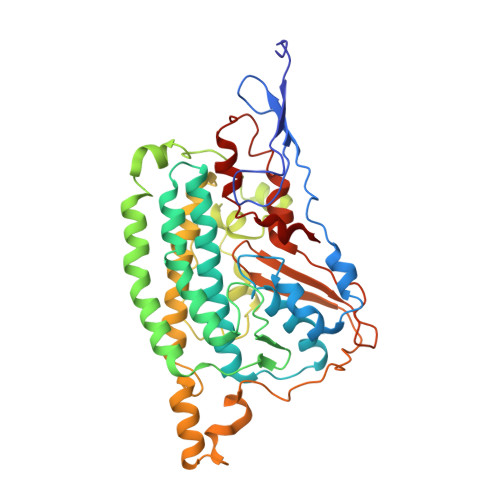
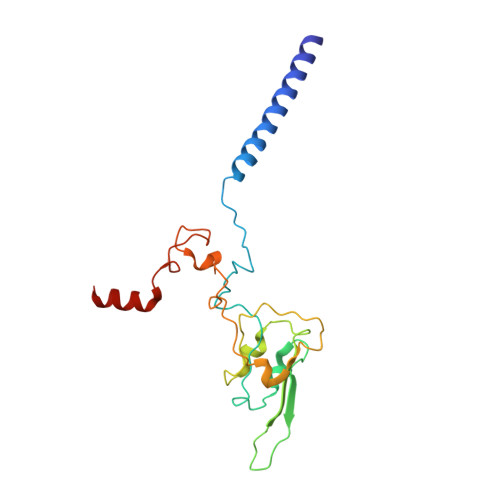
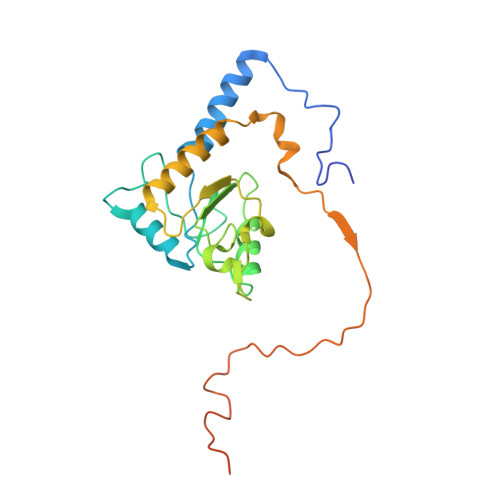
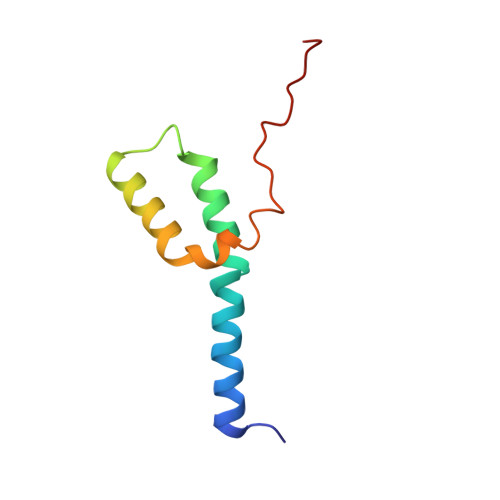
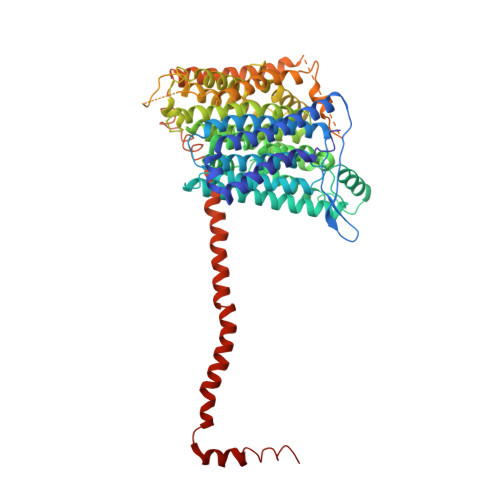
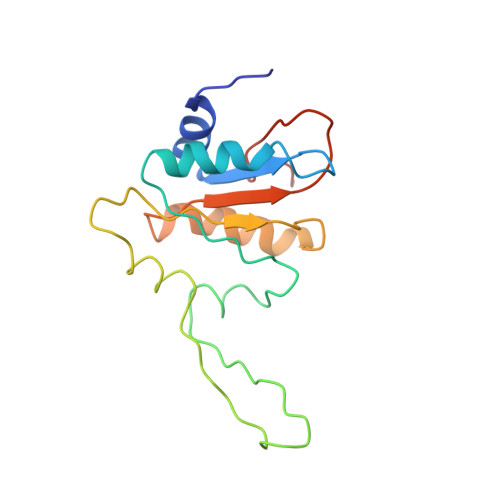
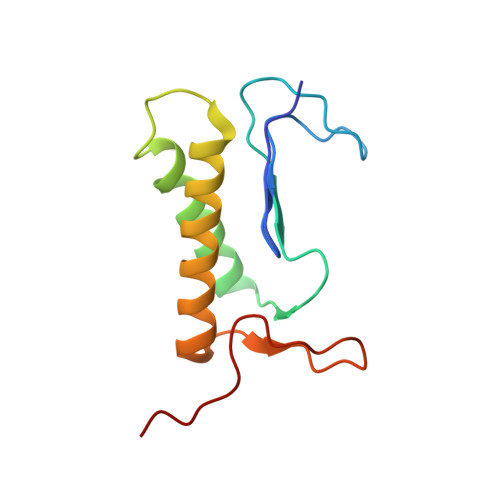
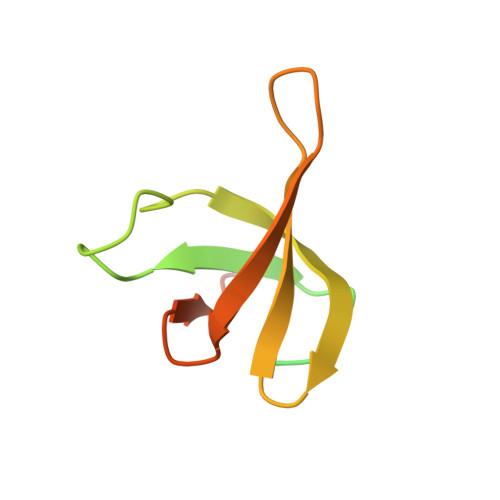
Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Schuller, J.M., Birrell, J.A., Tanaka, H., Konuma, T., Wulfhorst, H., Cox, N., Schuller, S.K., Thiemann, J., Lubitz, W., Setif, P., Ikegami, T., Engel, B.D., Kurisu, G., Nowaczyk, M.M.(2019) Science 363: 257-260
- PubMed: 30573545
- DOI: https://doi.org/10.1126/science.aau3613
- Primary Citation of Related Structures:
6A7K, 6HUM - PubMed Abstract:
Photosynthetic complex I enables cyclic electron flow around photosystem I, a regulatory mechanism for photosynthetic energy conversion. We report a 3.3-angstrom-resolution cryo-electron microscopy structure of photosynthetic complex I from the cyanobacterium Thermosynechococcus elongatus. The model reveals structural adaptations that facilitate binding and electron transfer from the photosynthetic electron carrier ferredoxin. By mimicking cyclic electron flow with isolated components in vitro, we demonstrate that ferredoxin directly mediates electron transfer between photosystem I and complex I, instead of using intermediates such as NADPH (the reduced form of nicotinamide adenine dinucleotide phosphate). A large rate constant for association of ferredoxin to complex I indicates efficient recognition, with the protein subunit NdhS being the key component in this process.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, 82152 Martinsried, Germany. janschu@biochem.mpg.de gkurisu@protein.osaka-u.ac.jp marc.m.nowaczyk@rub.de.
Organizational Affiliation: