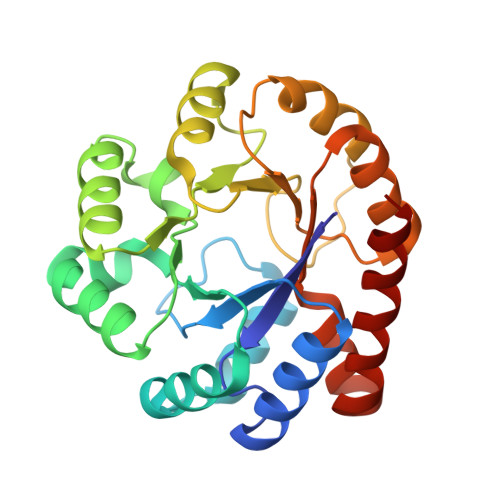
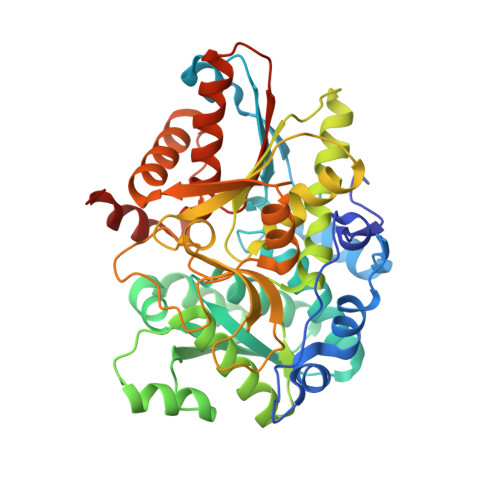
Evolutionary Morphing of Tryptophan Synthase: Functional Mechanisms for the Enzymatic Channeling of Indole.
Fleming, J.R., Schupfner, M., Busch, F., Basle, A., Ehrmann, A., Sterner, R., Mayans, O.(2018) J Mol Biology 430: 5066-5079
- PubMed: 30367843
- DOI: https://doi.org/10.1016/j.jmb.2018.10.013
- Primary Citation of Related Structures:
5N2P, 6HTE, 6HUL - PubMed Abstract:
Tryptophan synthase (TrpS) is a heterotetrameric αββα enzyme that exhibits complex substrate channeling and allosteric mechanisms and is a model system in enzymology. In this work, we characterize proposed early and late evolutionary states of TrpS and show that they have distinct quaternary structures caused by insertions-deletions of sequence segments (indels) in the β-subunit. Remarkably, indole hydrophobic channels that connect α and β active sites have re-emerged in both TrpS types, yet they follow different paths through the β-subunit fold. Also, both TrpS geometries activate the α-subunit through the rearrangement of loops flanking the active site. Our results link evolutionary sequence changes in the enzyme subunits with channeling and allostery in the TrpS enzymes. The findings demonstrate that indels allow protein quaternary architectures to escape "minima" in the evolutionary landscape, thereby overcoming the conservational constraints imposed by existing functional interfaces and being free to morph into new mechanistic enzymes.
- Department of Biology, University of Konstanz, 78457 Konstanz, Germany.
Organizational Affiliation: