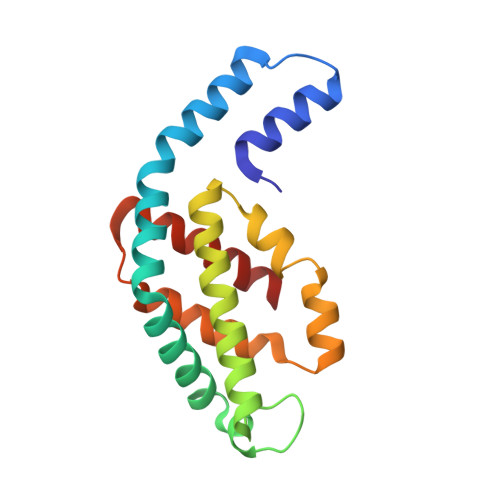
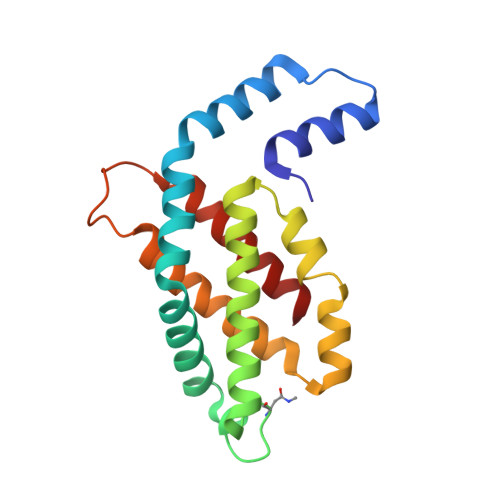
Crystal structure of phycocyanin from heterocyst-forming filamentous cyanobacterium Nostoc sp. WR13.
Patel, H.M., Roszak, A.W., Madamwar, D., Cogdell, R.J.(2019) Int J Biol Macromol 135: 62-68
- PubMed: 31121226
- DOI: https://doi.org/10.1016/j.ijbiomac.2019.05.099
- Primary Citation of Related Structures:
6HRN - PubMed Abstract:
Phycocyanin (PC) is the principal pigment protein in the light-harvesting antenna of cyanobacteria. Here the biochemical characterization and the 1.51 Å crystal structure of PC from cyanobacterium Nostoc sp. WR13 (Nst-PC) is reported. The P6 3 crystal lattice is composed of the minimal biological entities of Nst-PC, the (αβ) 3 trimeric rings. The structure has been refined to R factor 11.5% (R free 15.4%) using anisotropic atomic B factors. A phylogenetic study shows that the α and β chains of Nst-PC are significantly clustered in a distinct clade with Acaryochloris marina. The structure was examined to look for any significant differences between Nst-PC and PC from non-desert species. Only minor differences were found in the chromophore microenvironments. The tentative energy transfer pathways in Nst-PC were modeled based on simple structural considerations.
- P. G. Department of Biosciences, Satellite Campus, Vadtal Road, Sardar Patel University, Bakrol, 388315 Anand, Gujarat, India; Institute of Molecular Cell and Systems Biology, University of Glasgow, 120 University Place, Glasgow G12 8TA, UK. Electronic address: hiralpatel2387@yahoo.co.in.
Organizational Affiliation: