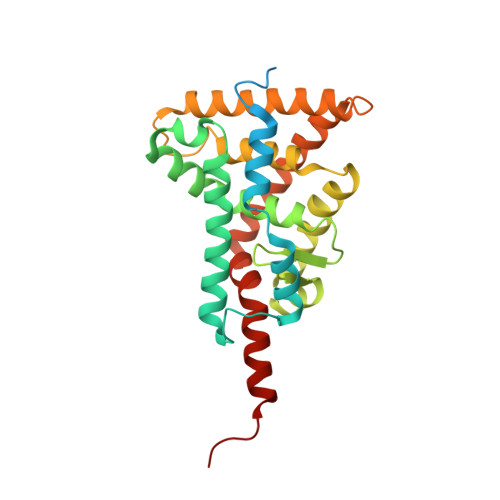
A revisited version of the apo structure of the ligand-binding domain of the human nuclear receptor retinoic X receptor alpha.
Eberhardt, J., McEwen, A.G., Bourguet, W., Moras, D., Dejaegere, A.(2019) Acta Crystallogr F Struct Biol Commun 75: 98-104
- PubMed: 30713160
- DOI: https://doi.org/10.1107/S2053230X18018022
- Primary Citation of Related Structures:
6HN6 - PubMed Abstract:
The retinoic X receptor (RXR) plays a crucial role in the superfamily of nuclear receptors (NRs) by acting as an obligatory partner of several nuclear receptors; its role as a transcription factor is thus critical in many signalling pathways, such as metabolism, cell development, differentiation and cellular death. The first published structure of the apo ligand-binding domain (LBD) of RXRα, which is still used as a reference today, contained inaccuracies. In the present work, these inaccuracies were corrected using modern crystallographic tools. The most important correction concerns the presence of a π-bulge in helix H7, which was originally built as a regular α-helix. The presence of several CHAPS molecules, which are visible for the first time in the electron-density map and which stabilize the H1-H3 loop, which contains helix H2, are also revealed. The apo RXR structure has played an essential role in deciphering the molecular mode of action of NR ligands and is still used in numerous biophysical studies. This refined structure should be used preferentially in the future in interpreting experiments as well as for modelling and structural dynamics studies of the apo RXRα LBD.
- Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France.
Organizational Affiliation: