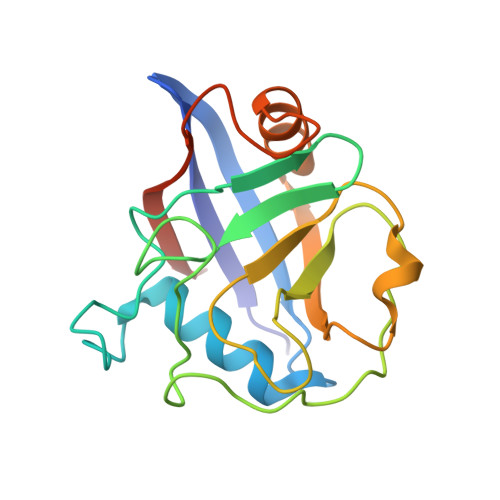
Enzyme activity and structural features of three single-domain phloem cyclophilins from Brassica napus.
Hanhart, P., Falke, S., Garbe, M., Rose, V., Thiess, M., Betzel, C., Kehr, J.(2019) Sci Rep 9: 9368-9368
- PubMed: 31249367
- DOI: https://doi.org/10.1038/s41598-019-45856-y
- Primary Citation of Related Structures:
6HMZ - PubMed Abstract:
Cyclophilins (CYPs) are a group of ubiquitous prolyl cis/trans isomerases (PPIases). It was shown that plants possess the most diverse CYP families and that these are abundant in the phloem long-distance translocation stream. Since phloem exudate showed PPIase activity, three single-domain CYPs that occur in phloem samples from Brassica napus were characterised on functional and structural levels. It could be shown that they exhibit isomerase activity and that this activity is controlled by a redox regulation mechanism, which has been postulated for divergent CYPs. The structure determination by small-angle X-ray scattering experiments revealed a conserved globular shape. In addition, the high-resolution crystal structure of BnCYP19-1 was resolved and refined to 2.0 Å resolution, and the active sites of related CYPs as well as substrate binding were modelled. The obtained data and results support the hypothesis that single domain phloem CYPs are active phloem PPIases that may function as chaperones.
- Universität Hamburg, Institute of Plant Science and Microbiology, Molecular Plant Genetics, Ohnhorststraße 18, 22609, Hamburg, Germany.
Organizational Affiliation: