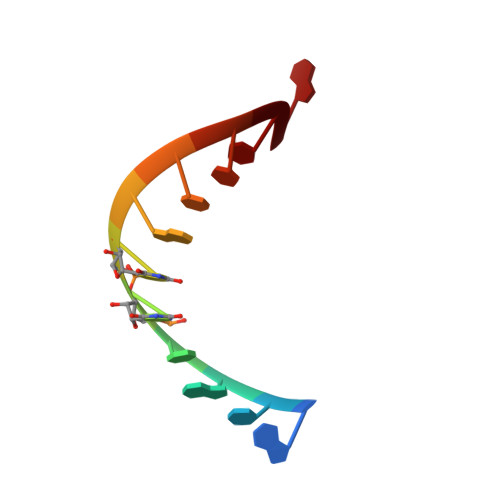
Structural basis of a small molecule targeting RNA for a specific splicing correction.
Campagne, S., Boigner, S., Rudisser, S., Moursy, A., Gillioz, L., Knorlein, A., Hall, J., Ratni, H., Clery, A., Allain, F.H.(2019) Nat Chem Biol 15: 1191-1198
- PubMed: 31636429
- DOI: https://doi.org/10.1038/s41589-019-0384-5
- Primary Citation of Related Structures:
6HMI, 6HMO - PubMed Abstract:
Splicing modifiers promoting SMN2 exon 7 inclusion have the potential to treat spinal muscular atrophy, the leading genetic cause of infantile death. These small molecules are SMN2 exon 7 selective and act during the early stages of spliceosome assembly. Here, we show at atomic resolution how the drug selectively promotes the recognition of the weak 5' splice site of SMN2 exon 7 by U1 snRNP. The solution structure of the RNA duplex formed following 5' splice site recognition in the presence of the splicing modifier revealed that the drug specifically stabilizes a bulged adenine at this exon-intron junction and converts the weak 5' splice site of SMN2 exon 7 into a stronger one. The small molecule acts as a specific splicing enhancer cooperatively with the splicing regulatory network. Our investigations uncovered a novel concept for gene-specific alternative splicing correction that we coined 5' splice site bulge repair.
- Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zurich, Zurich, Switzerland. sebastien.campagne@mol.biol.ethz.ch.
Organizational Affiliation: