Factor Inhibiting HIF (FIH) in complex with zinc, NOG and ASPP2 (970-992)
Leissing, T.M., Clifton, I.J., Lu, X., Schofield, C.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hypoxia-inducible factor 1-alpha inhibitor | 350 | Homo sapiens | Mutation(s): 0 Gene Names: HIF1AN, FIH1 EC: 1.14.11.30 (PDB Primary Data), 1.14.11 (PDB Primary Data) | 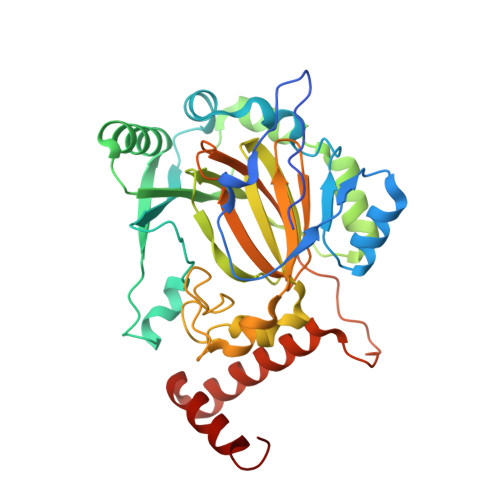 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9NWT6 (Homo sapiens) Explore Q9NWT6 Go to UniProtKB: Q9NWT6 | |||||
PHAROS: Q9NWT6 GTEx: ENSG00000166135 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9NWT6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Apoptosis-stimulating of p53 protein 2 | B [auth S] | 23 | Homo sapiens | Mutation(s): 0 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13625 (Homo sapiens) Explore Q13625 Go to UniProtKB: Q13625 | |||||
PHAROS: Q13625 GTEx: ENSG00000143514 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13625 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| OGA Query on OGA | N [auth A] | N-OXALYLGLYCINE C4 H5 N O5 BIMZLRFONYSTPT-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | C [auth A] D [auth A] E [auth A] F [auth A] G [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | I [auth A], J [auth A], K [auth A], L [auth A], M [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | O [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 86.02 | α = 90 |
| b = 86.02 | β = 90 |
| c = 148.508 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Engineering and Physical Sciences Research Council | United Kingdom | P/G03706X/1 |