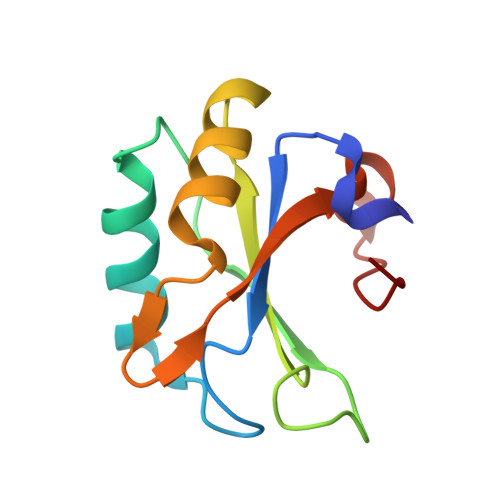
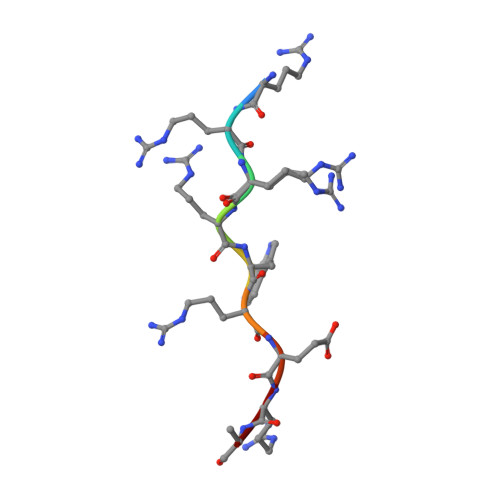
Modulation of HIV-1 gene expression by binding of a ULM motif in the Rev protein to UHM-containing splicing factors.
Pabis, M., Corsini, L., Vincendeau, M., Tripsianes, K., Gibson, T.J., Brack-Werner, R., Sattler, M.(2019) Nucleic Acids Res 47: 4859-4871
- PubMed: 30892606
- DOI: https://doi.org/10.1093/nar/gkz185
- Primary Citation of Related Structures:
6HIP - PubMed Abstract:
The HIV-1 protein Rev is essential for virus replication and ensures the expression of partially spliced and unspliced transcripts. We identified a ULM (UHM ligand motif) motif in the Arginine-Rich Motif (ARM) of the Rev protein. ULMs (UHM ligand motif) mediate protein interactions during spliceosome assembly by binding to UHM (U2AF homology motifs) domains. Using NMR, biophysical methods and crystallography we show that the Rev ULM binds to the UHMs of U2AF65 and SPF45. The highly conserved Trp45 in the Rev ULM is crucial for UHM binding in vitro, for Rev co-precipitation with U2AF65 in human cells and for proper processing of HIV transcripts. Thus, Rev-ULM interactions with UHM splicing factors contribute to the regulation of HIV-1 transcript processing, also at the splicing level. The Rev ULM is an example of viral mimicry of host short linear motifs that enables the virus to interfere with the host molecular machinery.
- Institute of Structural Biology, Helmholtz Zentrum München, Neuherberg 85 764, Germany.
Organizational Affiliation: