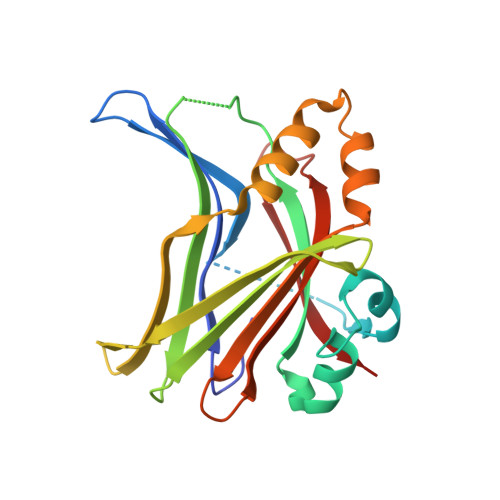
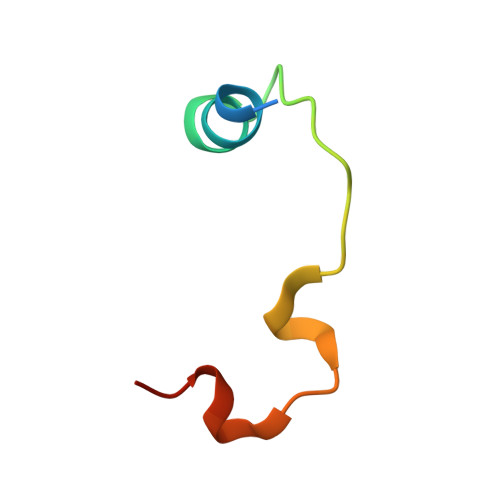
Molecular and structural characterization of a TEAD mutation at the origin of Sveinsson's chorioretinal atrophy.
Bokhovchuk, F., Mesrouze, Y., Izaac, A., Meyerhofer, M., Zimmermann, C., Fontana, P., Schmelzle, T., Erdmann, D., Furet, P., Kallen, J., Chene, P.(2019) FEBS J 286: 2381-2398
- PubMed: 30903741
- DOI: https://doi.org/10.1111/febs.14817
- Primary Citation of Related Structures:
6HIK, 6HIL - PubMed Abstract:
Four TEAD transcription factors (TEAD1-4) mediate the signalling output of the Hippo pathway that controls organ size in humans. TEAD transcriptional activity is regulated via interactions with the YAP, TAZ and VGLL proteins. A mutation in the TEAD1 gene, Tyr421His, has been identified in patients suffering from Sveinsson's chorioretinal atrophy (SCA), an autosomal dominant eye disease. This mutation prevents the YAP/TAZ:TEAD1 interaction. In this study, we measure the affinity of YAP, TAZ and VGLL1 for the four human TEADs and find that they have a similar affinity for all TEADs. We quantitate the effect of the mutation found in SCA patients and show that it destabilizes the YAP/TAZ:TEAD interaction by about 3 kcal·mol -1 . We determine the structure of YAP in complex with this mutant form of TEAD and show that the histidine residue adopts different conformations at the binding interface. The presence of this flexible residue induces the destabilization of several H-bonds and the loss of van der Waals contacts, which explains why the Tyr421His TEAD 1 mutation has such a large destabilizing effect on the formation of the YAP:TEAD complex. DATABASE: The crystallographic data have been deposited at the RSCB Protein Data Bank (PDB, www.pdb.org) with the access codes: 6HIL (wt YAP :Tyr421His TEAD1 ), 6HIK (wt YAP :Tyr429His TEAD4 ).
- Disease Area Oncology, Novartis Institutes for Biomedical Research, Basel, Switzerland.
Organizational Affiliation: