Structural and biochemical evaluation of Ceratitis capitata odorant-binding protein 22 affinity for odorants involved in intersex communication.
Falchetto, M., Ciossani, G., Scolari, F., Di Cosimo, A., Nenci, S., Field, L.M., Mattevi, A., Zhou, J.J., Gasperi, G., Forneris, F.(2019) Insect Mol Biol 28: 431-443
- PubMed: 30548711
- DOI: https://doi.org/10.1111/imb.12559
- Primary Citation of Related Structures:
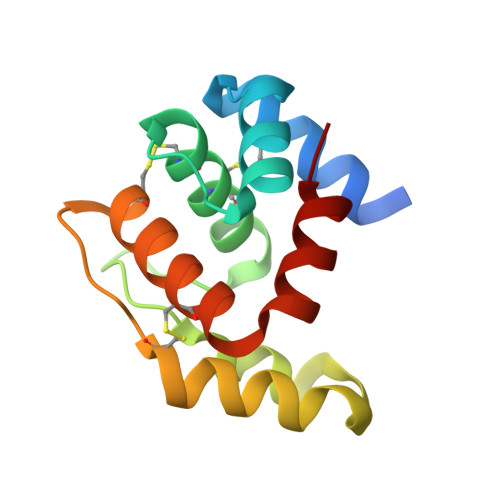
6HHE - PubMed Abstract:
In insects, odorant-binding proteins (OBPs) connect the peripheral sensory system to receptors of olfactory organs. Medfly Ceratitis capitata CcapObp22 shows 37% identity and close phylogenetic affinities with Drosophila melanogaster OBP69a/pheromone-binding protein related protein 1. The CcapObp22 gene is transcribed in the antennae and maxillary palps, suggesting an active role in olfaction. Here, we recombinantly produced CcapObp22, obtaining a 13.5 kDa protein capable of binding multiple strongly hydrophobic terpene compounds, including medfly male pheromone components. The highest binding affinity [half maximal effective concentration (EC50) = 0.48 µM] was to (E,E)-α-farnesene, one of the most abundant compounds in the male pheromone blend. This odorant was used in cocrystallization experiments, yielding the structure of CcapOBP22. The monomeric structure shows the typical OBP folding, constituted by six α-helical elements interconnected by three disulphide bridges. A C-terminal seventh α-helix constitutes the wall of a deep, L-shaped hydrophobic cavity. Analysis of the electron density in this cavity suggested trapping of farnesene in the crystal structure, although with partial occupancy. Superposition of the CcapOBP22 structure with related seven-helical OBPs highlights striking similarity in the organization of the C-terminal segment of these proteins. Collectively, our molecular and physiological data on medfly CcapOBP22 suggest its involvement in intersex olfactory communication.
- Department of Biology and Biotechnology "L. Spallanzani", University of Pavia, Pavia, Italy.
Organizational Affiliation: