Total Synthesis of Covalent Cysteine Protease Inhibitor N-Desmethyl Thalassospiramide C and Crystallographic Evidence for Its Mode of Action.
Fournier, J., Chen, K., Mailyan, A.K., Jackson, J.J., Buckman, B.O., Emayan, K., Yuan, S., Rajagopalan, R., Misialek, S., Adler, M., Blaesse, M., Griessner, A., Zakarian, A.(2019) Org Lett 21: 508-512
- PubMed: 30628449
- DOI: https://doi.org/10.1021/acs.orglett.8b03821
- Primary Citation of Related Structures:
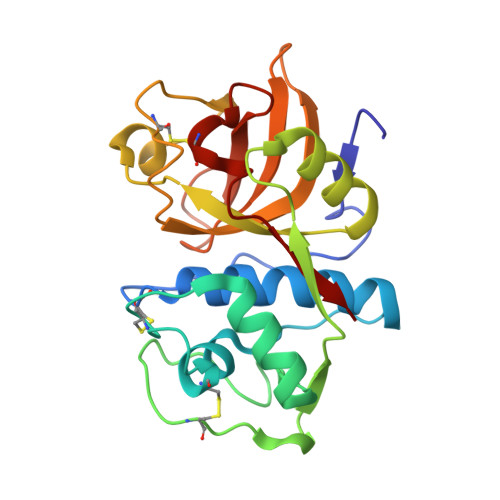
6HGY - PubMed Abstract:
A total synthesis of N-desmethyl thalassospiramide C, a unique strained macrocyclic proteobacterial depsipeptide, enabled a detailed crystallographic study of its covalent complex with cathepsin K, a member of a medicinally important family of cysteine proteases. The study provides support for the mechanism of action, and the insight gained can be used for structure-based drug design targeting these calpain proteases.
- Department of Chemistry and Biochemistry , University of California , Santa Barbara , California 93111 , United States.
Organizational Affiliation: