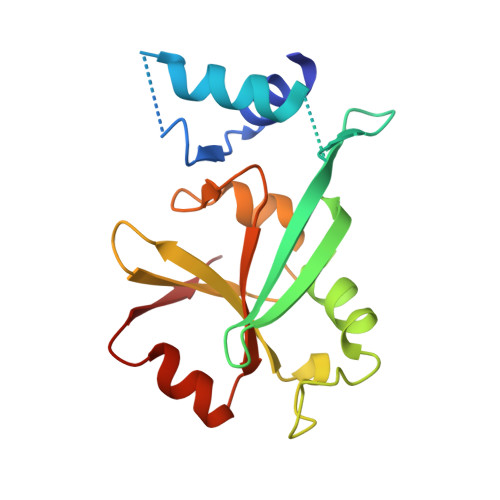
The Co-chaperone Cns1 and the Recruiter Protein Hgh1 Link Hsp90 to Translation Elongation via Chaperoning Elongation Factor 2.
Schopf, F.H., Huber, E.M., Dodt, C., Lopez, A., Biebl, M.M., Rutz, D.A., Muhlhofer, M., Richter, G., Madl, T., Sattler, M., Groll, M., Buchner, J.(2019) Mol Cell 74: 73-87.e8
- PubMed: 30876805
- DOI: https://doi.org/10.1016/j.molcel.2019.02.011
- Primary Citation of Related Structures:
6HFM, 6HFO, 6HFT - PubMed Abstract:
The Hsp90 chaperone machinery in eukaryotes comprises a number of distinct accessory factors. Cns1 is one of the few essential co-chaperones in yeast, but its structure and function remained unknown. Here, we report the X-ray structure of the Cns1 fold and NMR studies on the partly disordered, essential segment of the protein. We demonstrate that Cns1 is important for maintaining translation elongation, specifically chaperoning the elongation factor eEF2. In this context, Cns1 interacts with the novel co-factor Hgh1 and forms a quaternary complex together with eEF2 and Hsp90. The in vivo folding and solubility of eEF2 depend on the presence of these proteins. Chaperoning of eEF2 by Cns1 is essential for yeast viability and requires a defined subset of the Hsp90 machinery as well as the identified eEF2 recruiting factor Hgh1.
- Center for Integrated Protein Science at the Department Chemie, Technische Universität München, Lichtenbergstrasse 4, 85748 Garching, Germany.
Organizational Affiliation: