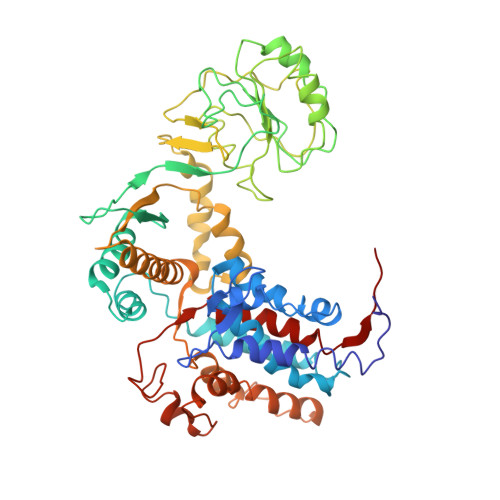
Cryo-EM reveals an asymmetry in a novel single-ring viral chaperonin.
Stanishneva-Konovalova, T.B., Semenyuk, P.I., Kurochkina, L.P., Pichkur, E.B., Vasilyev, A.L., Kovalchuk, M.V., Kirpichnikov, M.P., Sokolova, O.S.(2020) J Struct Biol 209: 107439-107439
- PubMed: 31870903
- DOI: https://doi.org/10.1016/j.jsb.2019.107439
- Primary Citation of Related Structures:
6HDD - PubMed Abstract:
Chaperonins are ubiquitously present protein complexes, which assist the proper folding of newly synthesized proteins and prevent aggregation of denatured proteins in an ATP-dependent manner. They are classified into group I (bacterial, mitochondrial, chloroplast chaperonins) and group II (archaeal and eukaryotic cytosolic variants). However, both of these groups do not include recently discovered viral chaperonins. Here, we solved the symmetry-free cryo-EM structures of a single-ring chaperonin encoded by the gene 246 of bacteriophage OBP Pseudomonas fluorescens, in the nucleotide-free, ATPγS-, and ADP-bound states, with resolutions of 4.3 Å, 5.0 Å, and 6 Å, respectively. The structure of OBP chaperonin reveals a unique subunit arrangement, with three pairs of subunits and one unpaired subunit. Each pair combines subunits in two possible conformations, differing in nucleotide-binding affinity. The binding of nucleotides results in the increase of subunits' conformational variability. Due to its unique structural and functional features, OBP chaperonin can represent a new group.
- Department of Bioengineering, Faculty of Biology, Lomonosov Moscow State University, Leninskie Gory 1, Bld 12, Moscow 119991, Russia.
Organizational Affiliation: