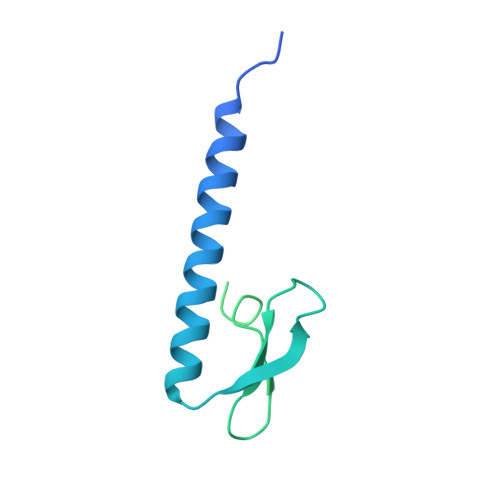
Sjogren syndrome/scleroderma autoantigen 1 is a direct Tankyrase binding partner in cancer cells.
Perdreau-Dahl, H., Progida, C., Barfeld, S.J., Guldsten, H., Thiede, B., Arntzen, M., Bakke, O., Mills, I.G., Krauss, S., Morth, J.P.(2020) Commun Biol 3: 123-123
- PubMed: 32170109
- DOI: https://doi.org/10.1038/s42003-020-0851-2
- Primary Citation of Related Structures:
6HCZ - PubMed Abstract:
Sjögren syndrome/scleroderma autoantigen 1 (SSSCA1) was first described as an auto-antigen over-expressed in Sjögren's syndrome and in scleroderma patients. SSSCA1 has been linked to mitosis and centromere association and as a potential marker candidate in diverse solid cancers. Here we characterize SSSCA1 for the first time, to our knowledge, at the molecular, structural and subcellular level. We have determined the crystal structure of a zinc finger fold, a zinc ribbon domain type 2 (ZNRD2), at 2.3 Å resolution. We show that the C-terminal domain serves a dual function as it both behaves as the interaction site to Tankyrase 1 (TNKS1) and as a nuclear export signal. We identify TNKS1 as a direct binding partner of SSSCA1, map the binding site to TNKS1 ankyrin repeat cluster 2 (ARC2) and thus define a new binding sequence. We experimentally verify and map a new nuclear export signal sequence in SSSCA1.
- Membrane Transport Group, Centre for Molecular Medicine Norway (NCMM), Nordic EMBL Partnership, University of Oslo, P.O. Box 1137 Blindern, 0318, Oslo, Norway.
Organizational Affiliation: