The role of RNA structure in translational regulation by L7Ae protein in archaea.
Huang, L., Ashraf, S., Lilley, D.M.J.(2019) RNA 25: 60-69
- PubMed: 30327333
- DOI: https://doi.org/10.1261/rna.068510.118
- Primary Citation of Related Structures:
6HCT - PubMed Abstract:
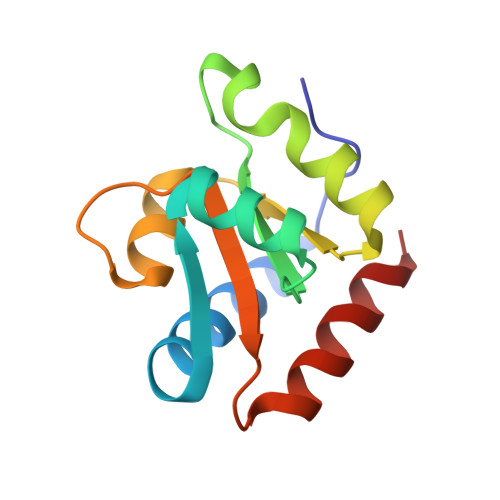
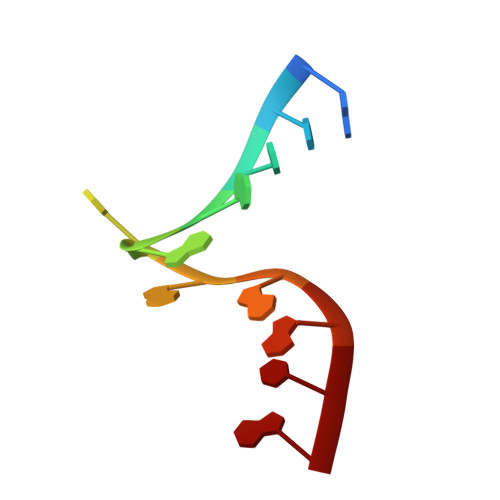
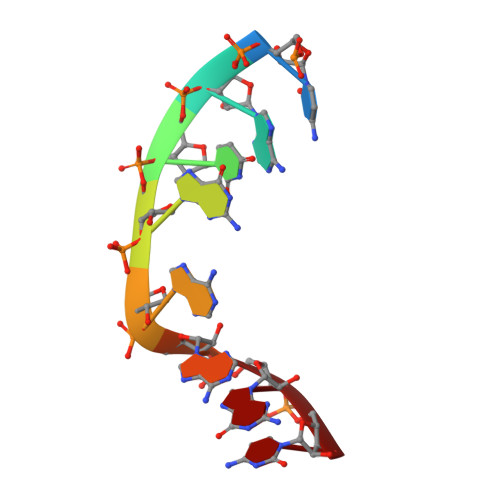
A recent study has shown that archaeal L7Ae binds to a putative k-turn structure in the 5'-leader of the mRNA of its structural gene to regulate translation. To function as a regulator, the RNA should be unstructured in the absence of protein, but it should adopt a k-turn-containing stem-loop on binding L7Ae. Sequence analysis of UTR sequences indicates that their k-turn elements will be unable to fold in the absence of L7Ae, and we have demonstrated this experimentally in solution using FRET for the Archaeoglobus fulgidus sequence. We have solved the X-ray crystal structure of the complex of the A. fulgidus RNA bound to its cognate L7Ae protein. The RNA adopts a standard k-turn conformation that is specifically recognized by the L7Ae protein, so stabilizing the stem-loop. In-line probing of the natural-sequence UTR shows that the RNA is unstructured in the absence of L7Ae binding, but folds on binding the protein such that the ribosome binding site is occluded. Thus, L7Ae regulates its own translation by switching the conformation of the RNA to alter accessibility.
- Cancer Research UK Nucleic Acid Structure Research Group, The University of Dundee, Dundee DD1 5EH, United Kingdom.
Organizational Affiliation: