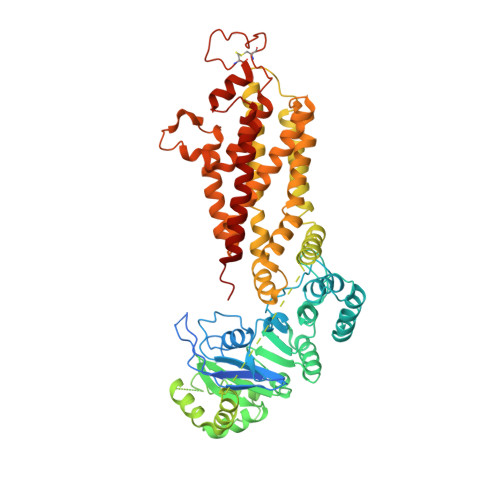
Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Manolaridis, I., Jackson, S.M., Taylor, N.M.I., Kowal, J., Stahlberg, H., Locher, K.P.(2018) Nature 563: 426-430
- PubMed: 30405239
- DOI: https://doi.org/10.1038/s41586-018-0680-3
- Primary Citation of Related Structures:
6HBU, 6HCO, 6HZM - PubMed Abstract:
ABCG2 is a transporter protein of the ATP-binding-cassette (ABC) family that is expressed in the plasma membrane in cells of various tissues and tissue barriers, including the blood-brain, blood-testis and maternal-fetal barriers 1-4 . Powered by ATP, it translocates endogenous substrates, affects the pharmacokinetics of many drugs and protects against a wide array of xenobiotics, including anti-cancer drugs 5-12 . Previous studies have revealed the architecture of ABCG2 and the structural basis of its inhibition by small molecules and antibodies 13,14 . However, the mechanisms of substrate recognition and ATP-driven transport are unknown. Here we present high-resolution cryo-electron microscopy (cryo-EM) structures of human ABCG2 in a substrate-bound pre-translocation state and an ATP-bound post-translocation state. For both structures, we used a mutant containing a glutamine replacing the catalytic glutamate (ABCG2 EQ ), which resulted in reduced ATPase and transport rates and facilitated conformational trapping for structural studies. In the substrate-bound state, a single molecule of estrone-3-sulfate (E 1 S) is bound in a central, hydrophobic and cytoplasm-facing cavity about halfway across the membrane. Only one molecule of E 1 S can bind in the observed binding mode. In the ATP-bound state, the substrate-binding cavity has collapsed while an external cavity has opened to the extracellular side of the membrane. The ATP-induced conformational changes include rigid-body shifts of the transmembrane domains, pivoting of the nucleotide-binding domains (NBDs), and a change in the relative orientation of the NBD subdomains. Mutagenesis and in vitro characterization of transport and ATPase activities demonstrate the roles of specific residues in substrate recognition, including a leucine residue that forms a 'plug' between the two cavities. Our results show how ABCG2 harnesses the energy of ATP binding to extrude E 1 S and other substrates, and suggest that the size and binding affinity of compounds are important for distinguishing substrates from inhibitors.
- Institute of Molecular Biology and Biophysics, Department of Biology, ETH Zurich, Switzerland.
Organizational Affiliation: