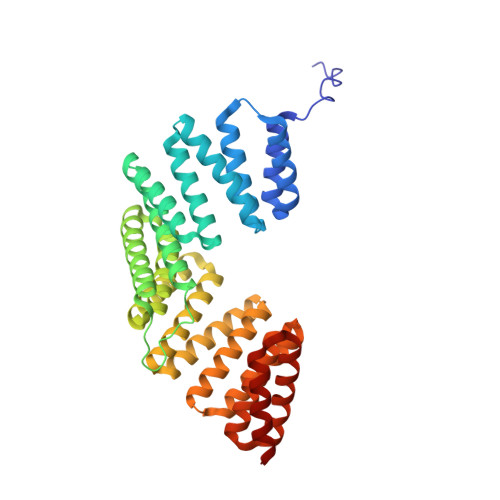
Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions.
Pirovano, L., Culurgioni, S., Carminati, M., Alfieri, A., Monzani, S., Cecatiello, V., Gaddoni, C., Rizzelli, F., Foadi, J., Pasqualato, S., Mapelli, M.(2019) Nat Commun 10: 2208-2208
- PubMed: 31101817
- DOI: https://doi.org/10.1038/s41467-019-09999-w
- Primary Citation of Related Structures:
6HC2 - PubMed Abstract:
Cortical force generators connect epithelial polarity sites with astral microtubules, allowing dynein movement to orient the mitotic spindle as astral microtubules depolymerize. Complexes of the LGN and NuMA proteins, fundamental components of force generators, are recruited to the cortex by Gαi-subunits of heterotrimeric G-proteins. They associate with dynein/dynactin and activate the motor activity pulling on astral microtubules. The architecture of cortical force generators is unknown. Here we report the crystal structure of NuMA:LGN hetero-hexamers, and unveil their role in promoting the assembly of active cortical dynein/dynactin motors that are required in orchestrating oriented divisions in polarized cells. Our work elucidates the basis for the structural organization of essential spindle orientation motors.
- IEO, European Institute of Oncology IRCCS, 20141, MILANO, Italy.
Organizational Affiliation: