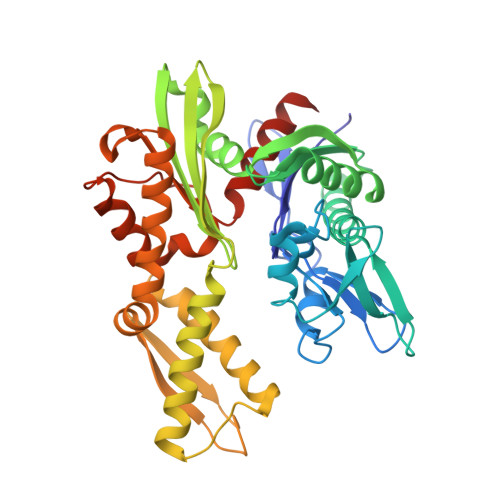
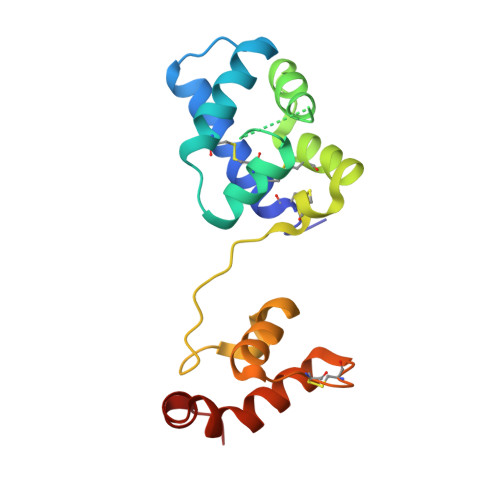
MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Yan, Y., Rato, C., Rohland, L., Preissler, S., Ron, D.(2019) Nat Commun 10: 541-541
- PubMed: 30710085
- DOI: https://doi.org/10.1038/s41467-019-08450-4
- Primary Citation of Related Structures:
6H9U, 6HA7, 6HAB - PubMed Abstract:
Despite its known role as a secreted neuroprotectant, much of the mesencephalic astrocyte-derived neurotrophic factor (MANF) is retained in the endoplasmic reticulum (ER) of producer cells. There, by unknown mechanisms, MANF plays a role in protein folding homeostasis in complex with the ER-localized Hsp70 chaperone BiP. Here we report that the SAF-A/B, Acinus, and PIAS (SAP) domain of MANF selectively associates with the nucleotide binding domain (NBD) of ADP-bound BiP. In crystal structures the SAP domain engages the cleft between NBD subdomains Ia and IIa, stabilizing the ADP-bound conformation and clashing with the interdomain linker that occupies this site in ATP-bound BiP. MANF inhibits both ADP release from BiP and ATP binding to BiP, and thereby client release. Cells lacking MANF have fewer ER stress-induced BiP-containing high molecular weight complexes. These findings suggest that MANF contributes to protein folding homeostasis as a nucleotide exchange inhibitor that stabilizes certain BiP-client complexes.
- Cambridge Institute for Medical Research, University of Cambridge, Cambridge, CB2 0XY, UK.
Organizational Affiliation: