1,1-Diheterocyclic Ethylenes Derived from Quinaldine and Carbazole as New Tubulin-Polymerization Inhibitors: Synthesis, Metabolism, and Biological Evaluation.
Naret, T., Khelifi, I., Provot, O., Bignon, J., Levaique, H., Dubois, J., Souce, M., Kasselouri, A., Deroussent, A., Paci, A., Varela, P.F., Gigant, B., Alami, M., Hamze, A.(2019) J Med Chem 62: 1902-1916
- PubMed: 30525602
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01386
- Primary Citation of Related Structures:
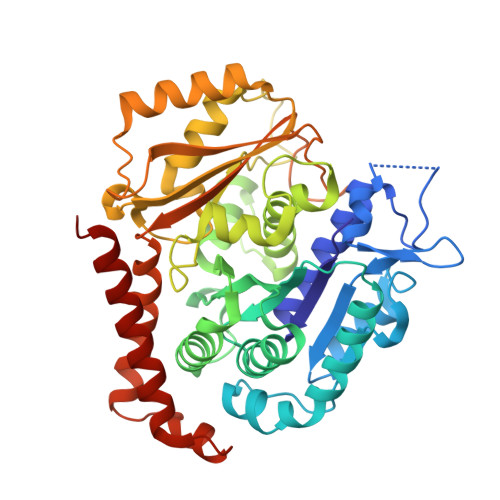
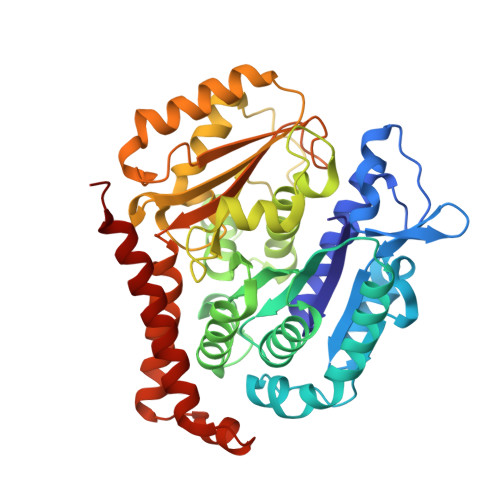
6H9B - PubMed Abstract:
We report the synthesis and metabolic and biological evaluation of a series of 17 novel heterocyclic derivatives of isocombretastatin-A4 (iso-CA-4) and their structure-activity relationships. Among these derivatives, the most active compound, 4f, inhibited the growth of a panel of seven cancer cell lines with an IC 50 in the low nanomolar range. In addition, 4f showed interesting activity against CA-4-resistant colon-carcinoma cells and multidrug-resistant leukemia cells. It also induced G 2 /M cell-cycle arrest. Structural data indicated binding of 4f to the colchicine site of tubulin, likely preventing the curved-to-straight tubulin structural changes that occur during microtubule assembly. Also, 4f disrupted the blood-vessel-like assembly formed by human umbilical-vein endothelial cells in vitro, suggesting its function as a vascular-disrupting agent. An in vitro metabolism study of 4f showed its high human-microsomal stability in comparison with that of iso-CA-4. The physicochemical properties of 4f may be conducive to CNS permeability, suggesting that this compound may be a possible candidate for the treatment of glioblastoma.
- BioCIS, Université Paris-Sud, CNRS, Équipe Labellisée Ligue Contre le Cancer , Université Paris-Saclay , F-92290 Châtenay-Malabry , France.
Organizational Affiliation: