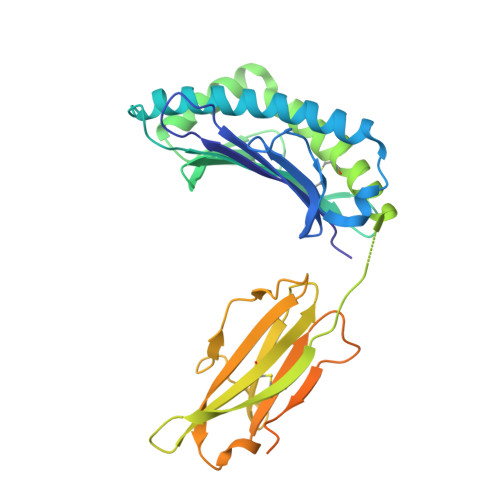
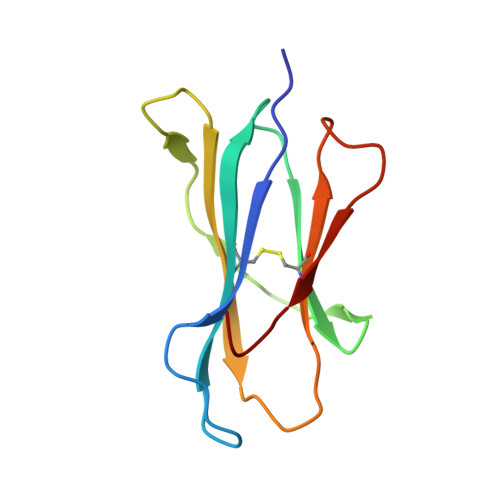
Crystal structures of H-2Db and H-2Dbm13 with cancer-associated Ad 10 peptide reveal that subtle changes in the peptide environment impact thermostability and alloreactivity
Han, X., Abualrous, E.T., Badia-Martines, D., Sandalova, T., Sun, R., van Hall, T., Ossendorp, F., Achour, A.To be published.