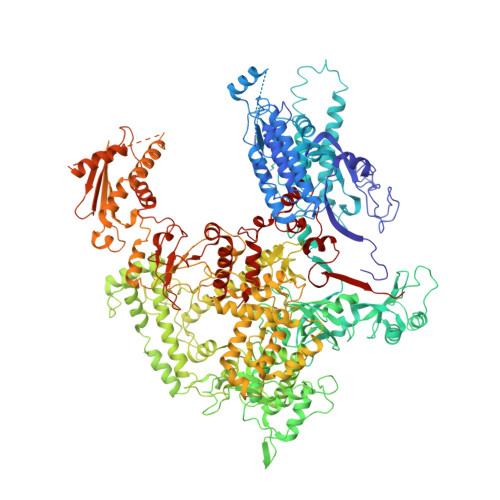
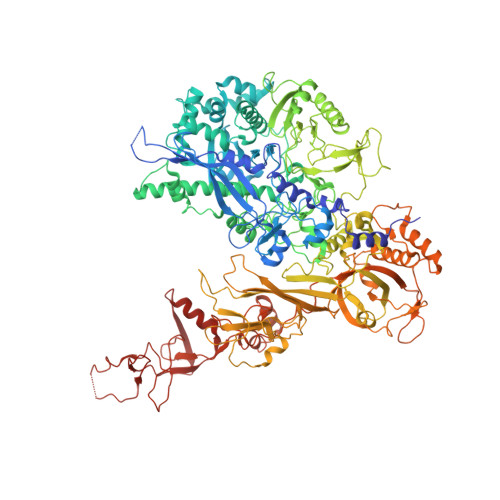
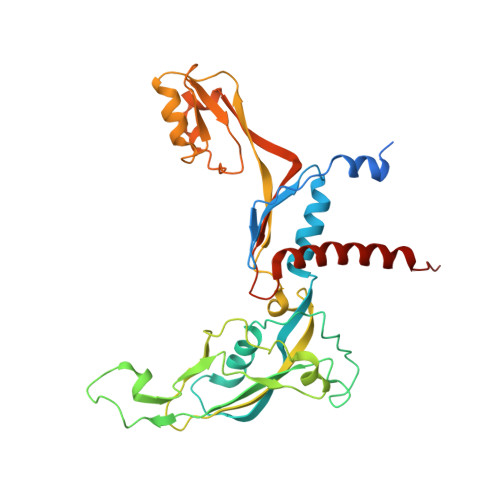
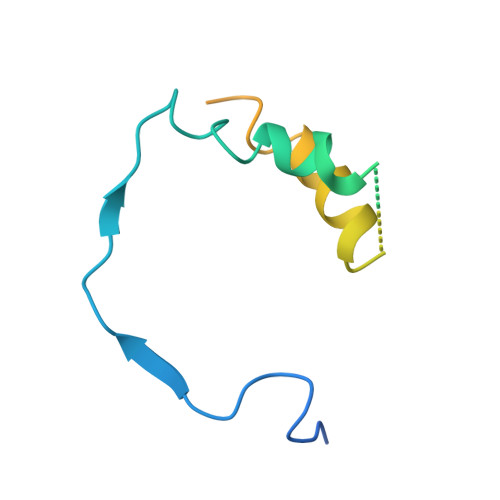
Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Sanz-Murillo, M., Xu, J., Belogurov, G.A., Calvo, O., Gil-Carton, D., Moreno-Morcillo, M., Wang, D., Fernandez-Tornero, C.(2018) Proc Natl Acad Sci U S A 115: 8972-8977
- PubMed: 30127008
- DOI: https://doi.org/10.1073/pnas.1802626115
- Primary Citation of Related Structures:
6H67, 6H68 - PubMed Abstract:
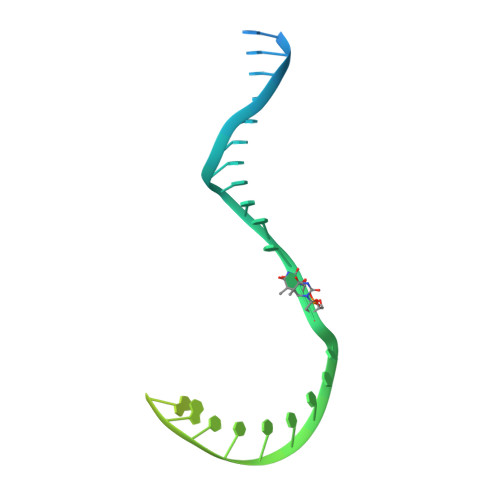
RNA polymerase I (Pol I) transcribes ribosomal DNA (rDNA) to produce the ribosomal RNA (rRNA) precursor, which accounts for up to 60% of the total transcriptional activity in growing cells. Pol I monitors rDNA integrity and influences cell survival, but little is known about how this enzyme processes UV-induced lesions. We report the electron cryomicroscopy structure of Pol I in an elongation complex containing a cyclobutane pyrimidine dimer (CPD) at a resolution of 3.6 Å. The structure shows that the lesion induces an early translocation intermediate exhibiting unique features. The bridge helix residue Arg1015 plays a major role in CPD-induced Pol I stalling, as confirmed by mutational analysis. These results, together with biochemical data presented here, reveal the molecular mechanism of Pol I stalling by CPD lesions, which is distinct from Pol II arrest by CPD lesions. Our findings open the avenue to unravel the molecular mechanisms underlying cell endurance to lesions on rDNA.
- Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Cientificas (CSIC), 28040 Madrid, Spain.
Organizational Affiliation: