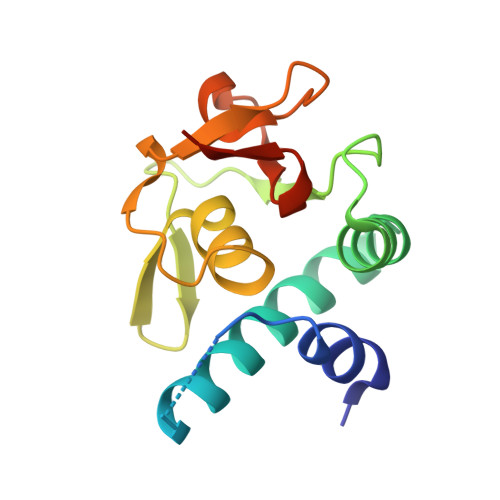
Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-beta signaling
Aragon, E., Wang, Q., Zou, Y., Morgani, S.M., Ruiz, L., Kaczmarska, Z., Su, J., Torner, C., Tian, L., Hu, J., Shu, W., Agrawal, S., Gomes, T., Marquez, J.A., Hadjantonakis, A.-K., Macias, M.J., Massague, J.(2020) Genes Dev