Water-soluble, stable and azide-reactive strained dialkynes for biocompatible double strain-promoted click chemistry.
Sharma, K., Strizhak, A.V., Fowler, E., Wang, X., Xu, W., Hatt Jensen, C., Wu, Y., Sore, H.F., Lau, Y.H., Hyvonen, M., Itzhaki, L.S., Spring, D.R.(2019) Org Biomol Chem 17: 8014-8018
- PubMed: 31418442
- DOI: https://doi.org/10.1039/c9ob01745c
- Primary Citation of Related Structures:
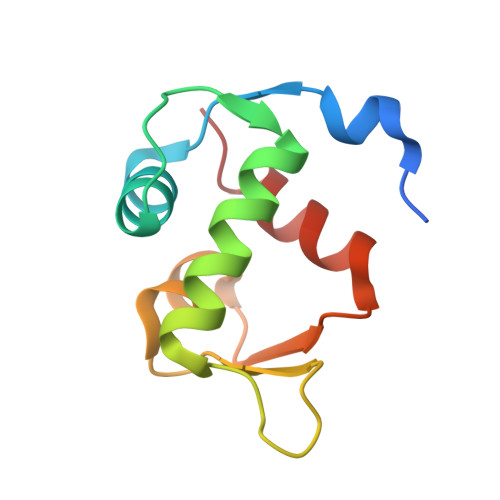
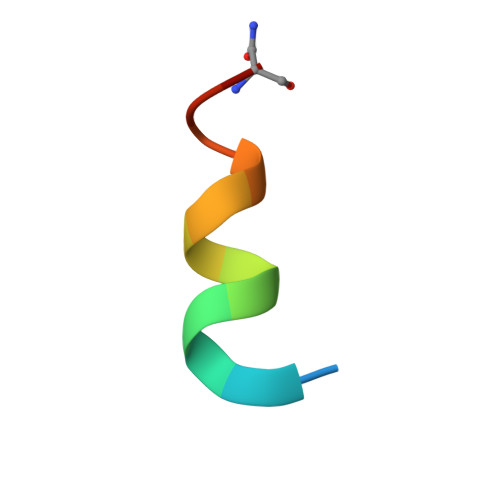
6H22 - PubMed Abstract:
The Sondheimer dialkyne is extensively used in double strain-promoted azide-alkyne cycloadditions. This reagent suffers with poor water-solubility and rapidly decomposes in aqueous solutions. This intrinsically limits its application in biological systems, and no effective solutions are currently available. Herein, we report the development of novel highly water-soluble, stable, and azide-reactive strained dialkyne reagents. To demonstrate their extensive utility, we applied our novel dialkynes to a double strain-promoted macrocyclisation strategy to generate functionalised p53-based stapled peptides for inhibiting the oncogenic p53-MDM2 interaction. These functionalised stapled peptides bind MDM2 with low nanomolar affinity and show p53 activation in a cellular environment. Overall, our highly soluble, stable and azide-reactive dialkynes offer significant advantages over the currently used Sondheimer dialkyne, and could be utilised for numerous biological applications.
- Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK.
Organizational Affiliation: