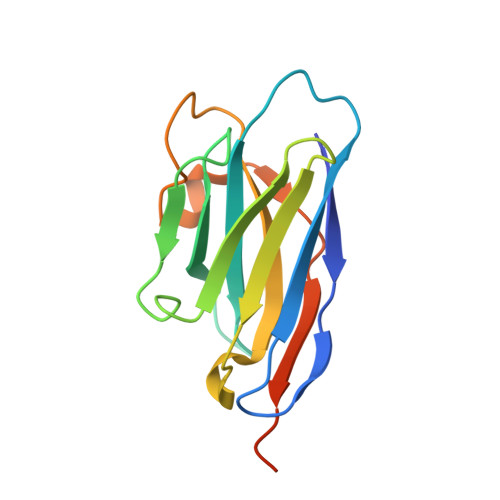
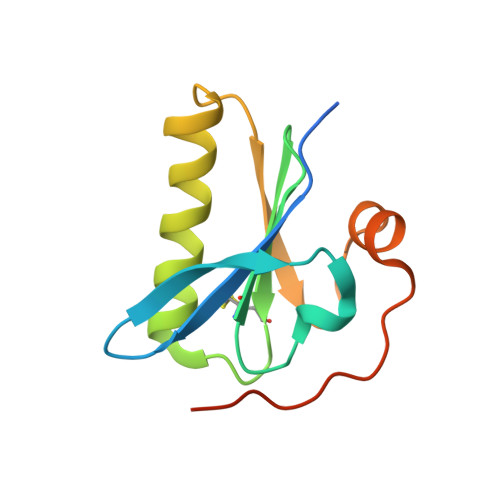
Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Giorgino, T., Mattioni, D., Hassan, A., Milani, M., Mastrangelo, E., Barbiroli, A., Verhelle, A., Gettemans, J., Barzago, M.M., Diomede, L., de Rosa, M.(2019) Biochim Biophys Acta Mol Basis Dis 1865: 648-660
- PubMed: 30625383
- DOI: https://doi.org/10.1016/j.bbadis.2019.01.010
- Primary Citation of Related Structures:
6H1F - PubMed Abstract:
AGel amyloidosis, formerly known as familial amyloidosis of the Finnish-type, is caused by pathological aggregation of proteolytic fragments of plasma gelsolin. So far, four mutations in the gelsolin gene have been reported as responsible for the disease. Although D187N is the first identified variant and the best characterized, its structure has been hitherto elusive. Exploiting a recently-developed nanobody targeting gelsolin, we were able to stabilize the G2 domain of the D187N protein and obtained, for the first time, its high-resolution crystal structure. In the nanobody-stabilized conformation, the main effect of the D187N substitution is the impairment of the calcium binding capability, leading to a destabilization of the C-terminal tail of G2. However, molecular dynamics simulations show that in the absence of the nanobody, D187N-mutated G2 further misfolds, ultimately exposing its hydrophobic core and the furin cleavage site. The nanobody's protective effect is based on the enhancement of the thermodynamic stability of different G2 mutants (D187N, G167R and N184K). In particular, the nanobody reduces the flexibility of dynamic stretches, and most notably decreases the conformational entropy of the C-terminal tail, otherwise stabilized by the presence of the Ca 2+ ion. A Caenorhabditis elegans-based assay was also applied to quantify the proteotoxic potential of the mutants and determine whether nanobody stabilization translates into a biologically relevant effect. Successful protection from G2 toxicity in vivo points to the use of C. elegans as a tool for investigating the mechanisms underlying AGel amyloidosis and rapidly screen new therapeutics.
- Istituto di Biofisica, Consiglio Nazionale delle Ricerche, Milano, Italy; Dipartimento di Bioscienze, Università degli Studi di Milano, Milano, Italy.
Organizational Affiliation: