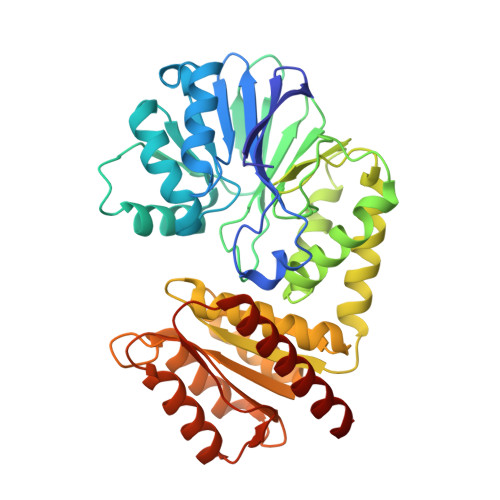
Analysis of a new flavodiiron core structural arrangement in Flv1-Delta FlR protein from Synechocystis sp. PCC6803.
Borges, P.T., Romao, C.V., Saraiva, L.M., Goncalves, V.L., Carrondo, M.A., Teixeira, M., Frazao, C.(2019) J Struct Biol 205: 91-102
- PubMed: 30447285
- DOI: https://doi.org/10.1016/j.jsb.2018.11.004
- Primary Citation of Related Structures:
6H0C, 6H0D - PubMed Abstract:
Flavodiiron proteins (FDPs) play key roles in biological response mechanisms against oxygen and/or nitric oxide; in particular they are present in oxygenic phototrophs (including cyanobacteria and gymnosperms). Two conserved domains define the core of this family of proteins: a N-terminal metallo-β-lactamase-like domain followed by a C-terminal flavodoxin-like one, containing the catalytic diiron centre and a FMN cofactor, respectively. Members of the FDP family may present extra modules in the C-terminus, and were classified into several classes according to their distribution and composition. The cyanobacterium Synechocystis sp. PCC6803 contains four Class C FDPs (Flv1-4) that include at the C-terminus an additional NAD(P)H:flavin oxidoreductase (FlR) domain. Two of them (Flv3 and Flv4) have the canonical diiron ligands (Class C, Type 1), while the other two (Flv1 and Flv2) present different residues in that region (Class C, Type 2). Most phototrophs, either Bacterial or Eukaryal, contain at least two FDP genes, each encoding for one of those two types. Crystals of the Flv1 two core domains (Flv1-ΔFlR), without the C-terminal NAD(P)H:flavin oxidoreductase extension, were obtained and the structure was determined. Its pseudo diiron site contains non-canonical basic and neutral residues, and showed anion moieties, instead. The presented structure revealed for the first time the structure of the two-domain core of a Class C-Type 2 FDP.
- ITQB NOVA, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Avenida da República, 2780-157 Oeiras, Portugal.
Organizational Affiliation: