Deep Structural Analysis of RPAP3 and PIH1D1, Two Components of the HSP90 Co-chaperone R2TP Complex.
Henri, J., Chagot, M.E., Bourguet, M., Abel, Y., Terral, G., Maurizy, C., Aigueperse, C., Georgescauld, F., Vandermoere, F., Saint-Fort, R., Behm-Ansmant, I., Charpentier, B., Pradet-Balade, B., Verheggen, C., Bertrand, E., Meyer, P., Cianferani, S., Manival, X., Quinternet, M.(2018) Structure 26: 1196-1209.e8
- PubMed: 30033218
- DOI: https://doi.org/10.1016/j.str.2018.06.002
- Primary Citation of Related Structures:
6FD7, 6FDP, 6FDT, 6GXZ - PubMed Abstract:
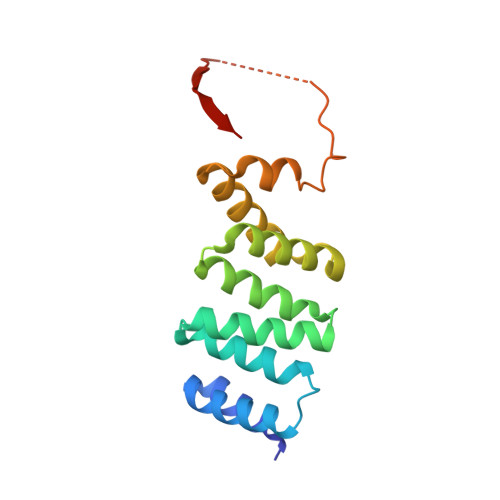
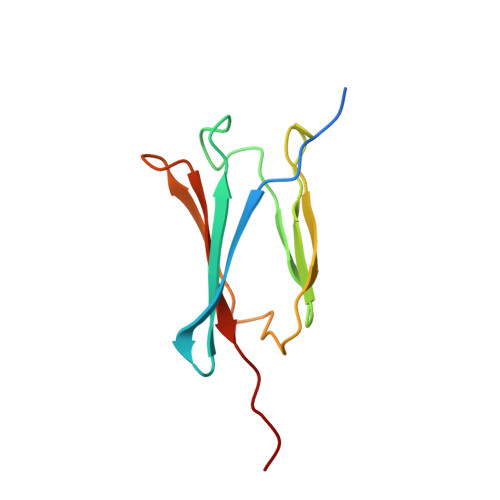
RPAP3 and PIH1D1 are part of the HSP90 co-chaperone R2TP complex involved in the assembly process of many molecular machines. In this study, we performed a deep structural investigation of the HSP binding abilities of the two TPR domains of RPAP3. We combined 3D NMR, non-denaturing MS, and ITC techniques with Y2H, IP-LUMIER, FRET, and ATPase activity assays and explain the fundamental role played by the second TPR domain of RPAP3 in the specific recruitment of HSP90. We also established the 3D structure of an RPAP3:PIH1D1 sub-complex demonstrating the need for a 34-residue insertion, specific of RPAP3 isoform 1, for the tight binding of PIH1D1. We also confirm the existence of a complex lacking PIH1D1 in human cells (R2T), which shows differential binding to certain clients. These results highlight similarities and differences between the yeast and human R2TP complexes, and document the diversification of this family of co-chaperone complexes in human.
- LBMCE, UMR 8226 CNRS Sorbonne Université, IBPC, 75005 Paris, France.
Organizational Affiliation: