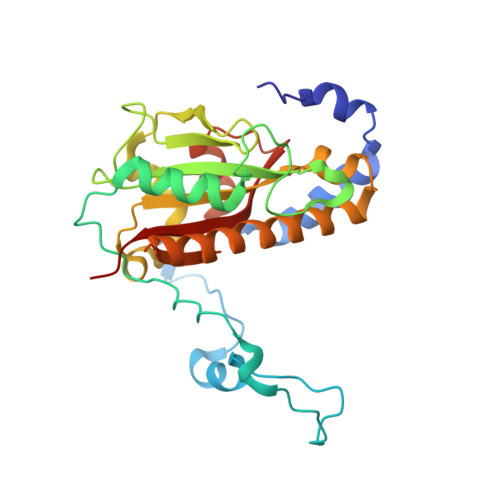
Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Ghilarov, D., Stevenson, C.E.M., Travin, D.Y., Piskunova, J., Serebryakova, M., Maxwell, A., Lawson, D.M., Severinov, K.(2019) Mol Cell 73: 749-762.e5
- PubMed: 30661981
- DOI: https://doi.org/10.1016/j.molcel.2018.11.032
- Primary Citation of Related Structures:
6GOS, 6GRG, 6GRH, 6GRI - PubMed Abstract:
The introduction of azole heterocycles into a peptide backbone is the principal step in the biosynthesis of numerous compounds with therapeutic potential. One of them is microcin B17, a bacterial topoisomerase inhibitor whose activity depends on the conversion of selected serine and cysteine residues of the precursor peptide to oxazoles and thiazoles by the McbBCD synthetase complex. Crystal structures of McbBCD reveal an octameric B 4 C 2 D 2 complex with two bound substrate peptides. Each McbB dimer clamps the N-terminal recognition sequence, while the C-terminal heterocycle of the modified peptide is trapped in the active site of McbC. The McbD and McbC active sites are distant from each other, which necessitates alternate shuttling of the peptide substrate between them, while remaining tethered to the McbB dimer. An atomic-level view of the azole synthetase is a starting point for deeper understanding and control of biosynthesis of a large group of ribosomally synthesized natural products.
- Centre for Life Sciences, Skolkovo Institute of Science and Technology, 143026 Moscow, Russia; Institute of Gene Biology of the Russian Academy of Sciences, 119334 Moscow, Russia; Malopolska Centre of Biotechnology, Jagiellonian University, 30-387 Cracow, Poland.
Organizational Affiliation: