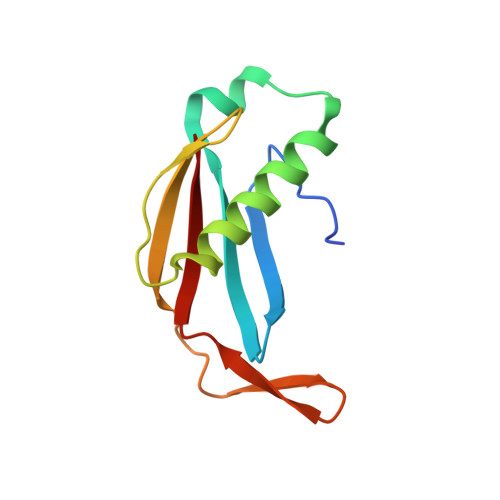
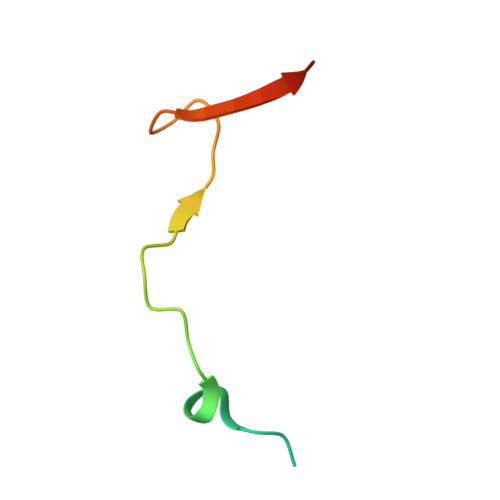
Structural basis of meiotic telomere attachment to the nuclear envelope by MAJIN-TERB2-TERB1.
Dunce, J.M., Milburn, A.E., Gurusaran, M., da Cruz, I., Sen, L.T., Benavente, R., Davies, O.R.(2018) Nat Commun 9: 5355-5355
- PubMed: 30559341
- DOI: https://doi.org/10.1038/s41467-018-07794-7
- Primary Citation of Related Structures:
6GNX, 6GNY - PubMed Abstract:
Meiotic chromosomes undergo rapid prophase movements, which are thought to facilitate the formation of inter-homologue recombination intermediates that underlie synapsis, crossing over and segregation. The meiotic telomere complex (MAJIN, TERB1, TERB2) tethers telomere ends to the nuclear envelope and transmits cytoskeletal forces via the LINC complex to drive these rapid movements. Here, we report the molecular architecture of the meiotic telomere complex through the crystal structure of MAJIN-TERB2, together with light and X-ray scattering studies of wider complexes. The MAJIN-TERB2 2:2 hetero-tetramer binds strongly to DNA and is tethered through long flexible linkers to the inner nuclear membrane and two TRF1-binding 1:1 TERB2-TERB1 complexes. Our complementary structured illumination microscopy studies and biochemical findings reveal a telomere attachment mechanism in which MAJIN-TERB2-TERB1 recruits telomere-bound TRF1, which is then displaced during pachytene, allowing MAJIN-TERB2-TERB1 to bind telomeric DNA and form a mature attachment plate.
- Institute for Cell and Molecular Biosciences, Faculty of Medical Sciences, Newcastle University, Framlington Place, Newcastle upon Tyne, NE2 4HH, UK.
Organizational Affiliation: