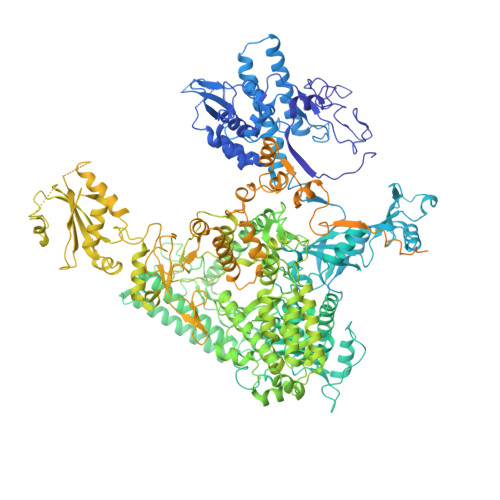
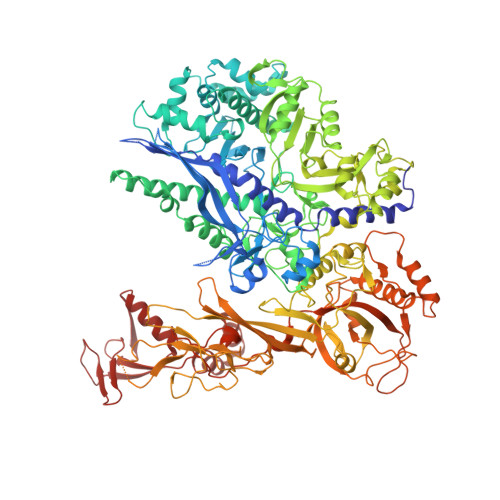
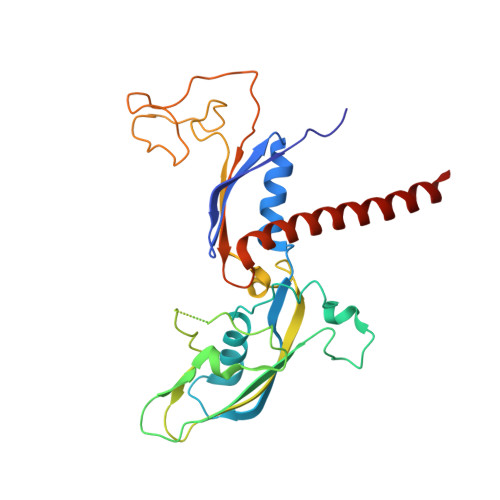
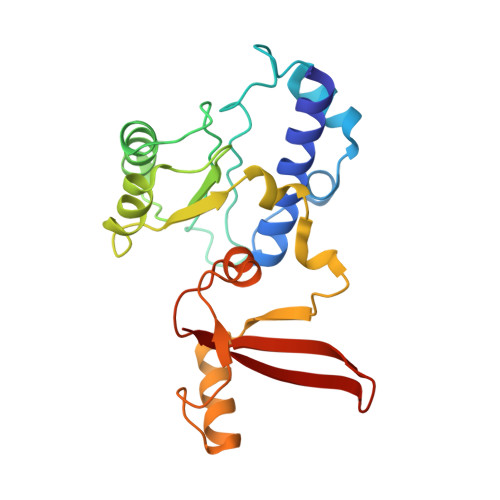
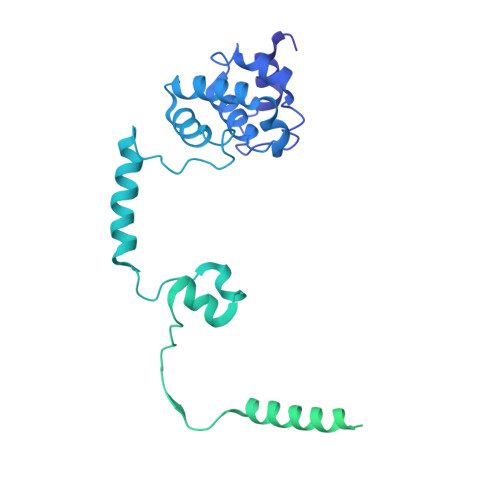
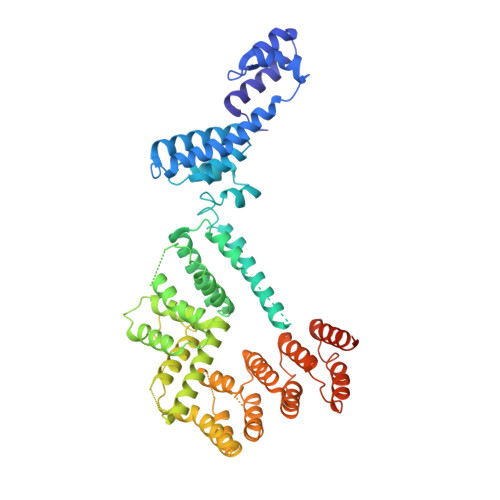
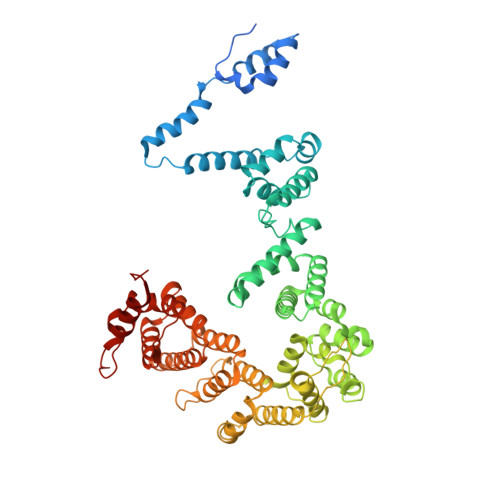
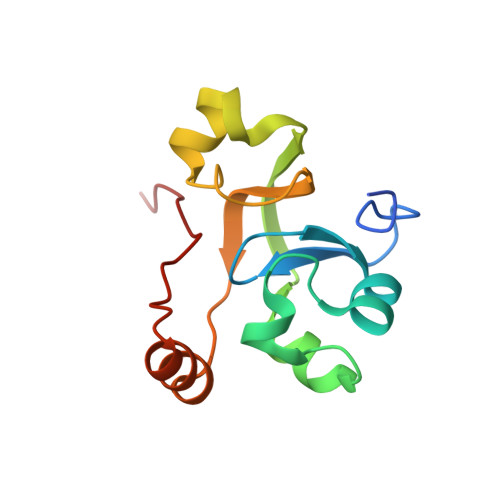
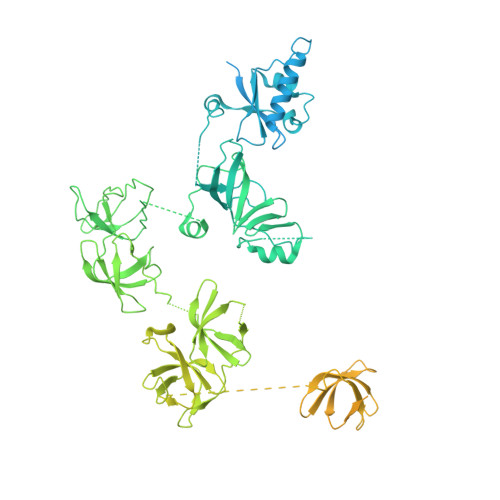
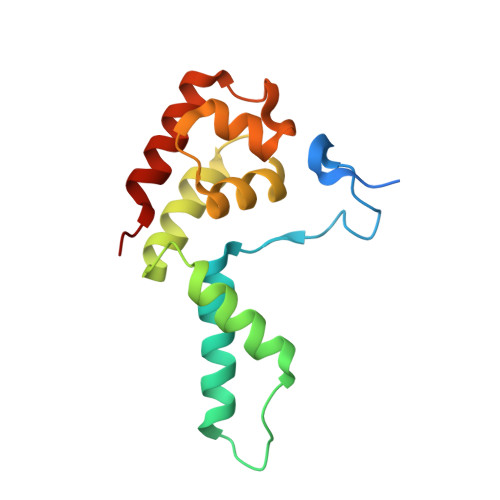
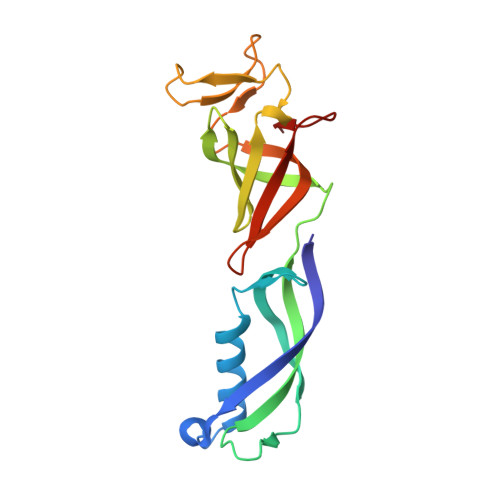
Structure of paused transcription complex Pol II-DSIF-NELF.
Vos, S.M., Farnung, L., Urlaub, H., Cramer, P.(2018) Nature 560: 601-606
- PubMed: 30135580
- DOI: https://doi.org/10.1038/s41586-018-0442-2
- Primary Citation of Related Structures:
6GML - PubMed Abstract:
Metazoan gene regulation often involves the pausing of RNA polymerase II (Pol II) in the promoter-proximal region. Paused Pol II is stabilized by the protein complexes DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF). Here we report the cryo-electron microscopy structure of a paused transcription elongation complex containing Sus scrofa Pol II and Homo sapiens DSIF and NELF at 3.2 Å resolution. The structure reveals a tilted DNA-RNA hybrid that impairs binding of the nucleoside triphosphate substrate. NELF binds the polymerase funnel, bridges two mobile polymerase modules, and contacts the trigger loop, thereby restraining Pol II mobility that is required for pause release. NELF prevents binding of the anti-pausing transcription elongation factor IIS (TFIIS). Additionally, NELF possesses two flexible 'tentacles' that can contact DSIF and exiting RNA. These results define the paused state of Pol II and provide the molecular basis for understanding the function of NELF during promoter-proximal gene regulation.
- Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Göttingen, Germany.
Organizational Affiliation: