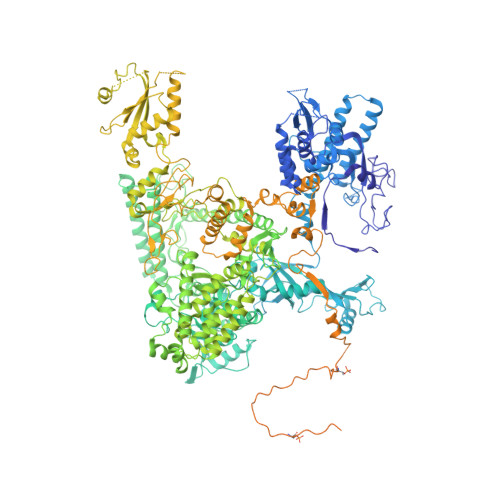
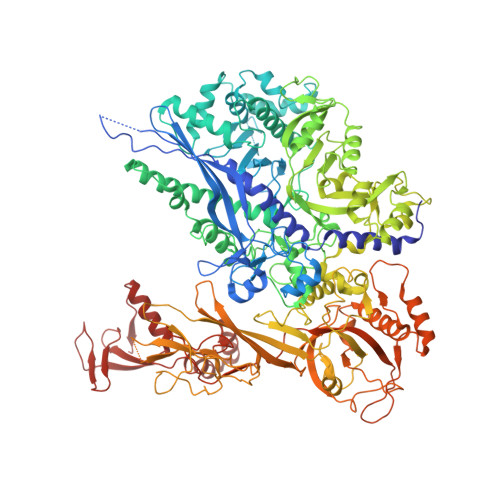
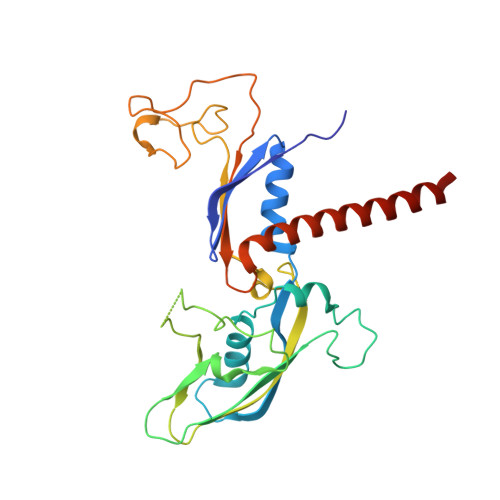
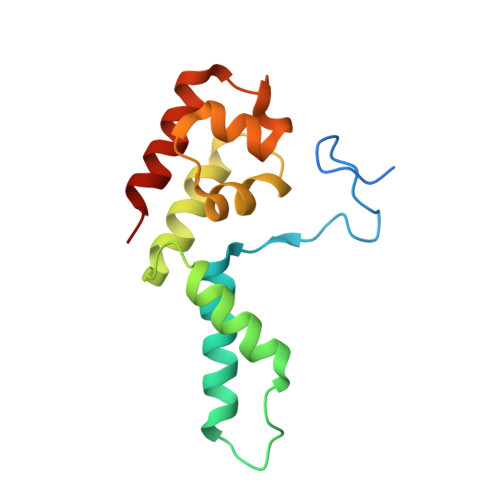
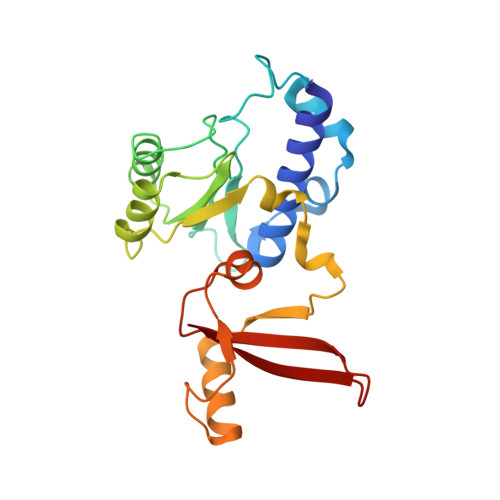
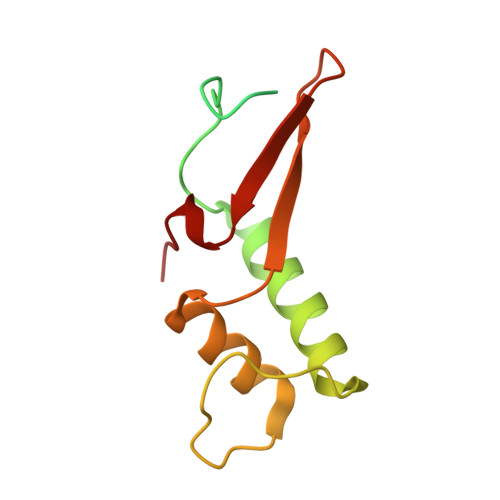
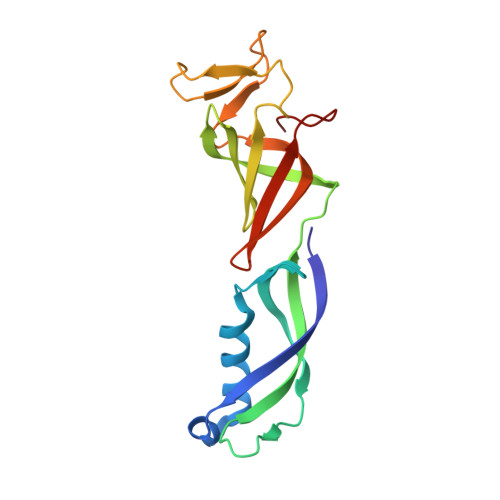
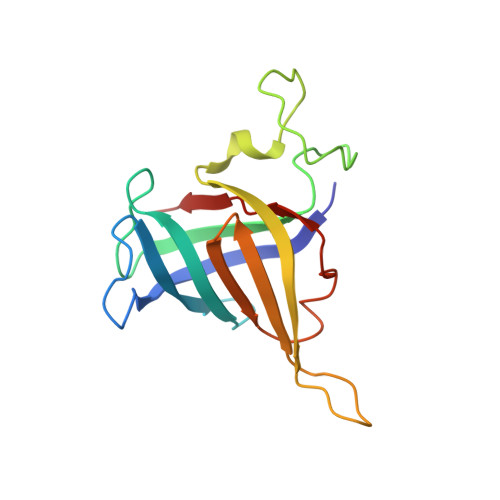
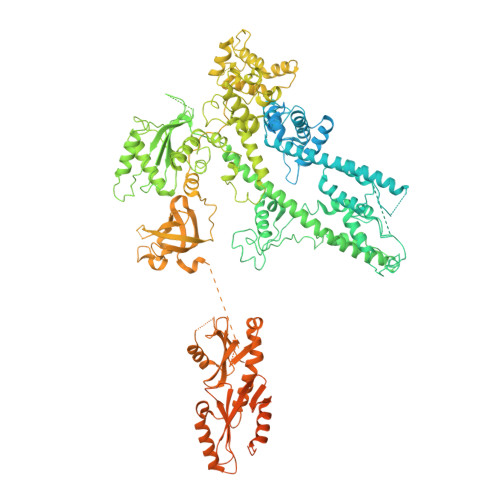
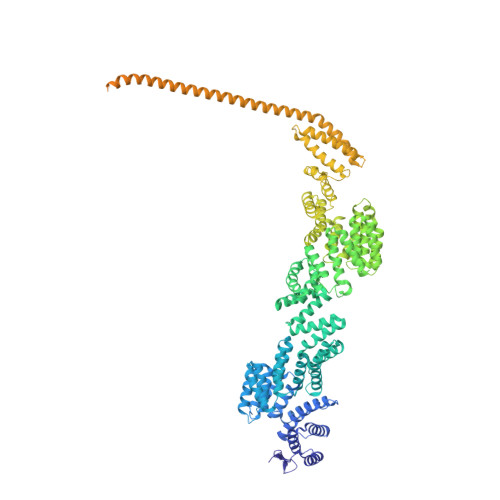
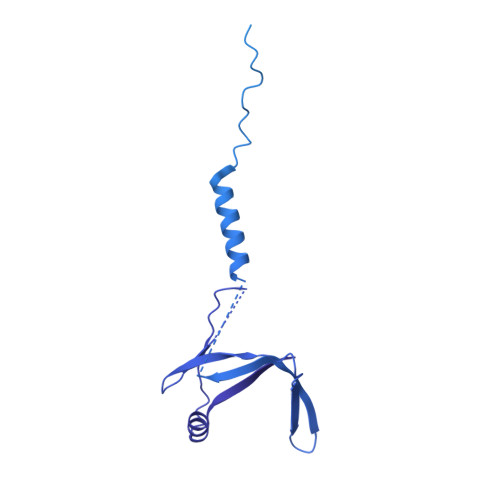
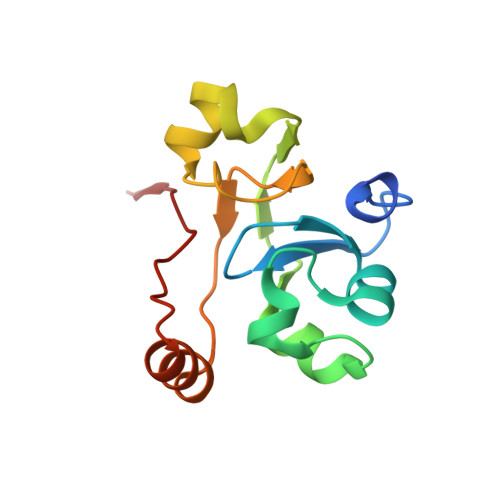
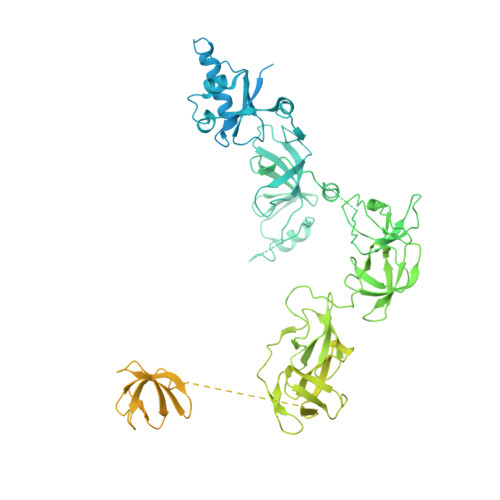
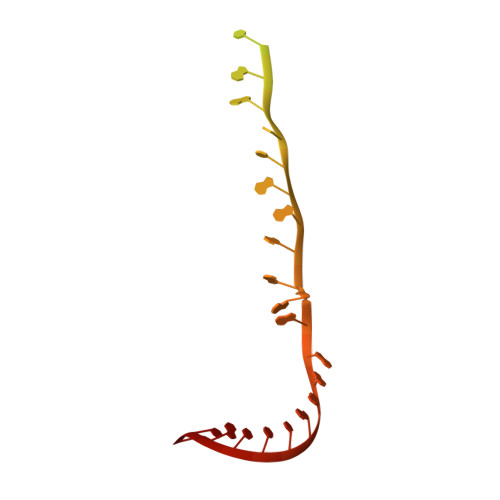
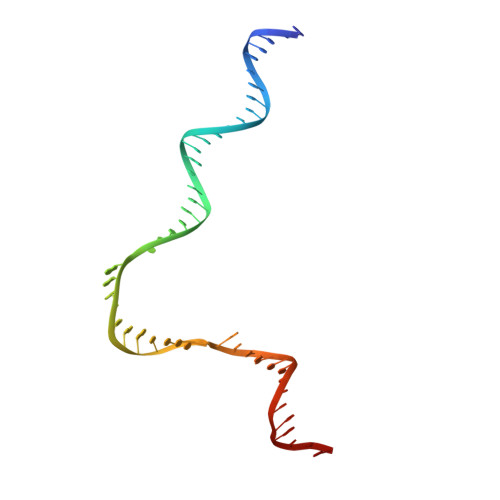
Structure of activated transcription complex Pol II-DSIF-PAF-SPT6.
Vos, S.M., Farnung, L., Boehning, M., Wigge, C., Linden, A., Urlaub, H., Cramer, P.(2018) Nature 560: 607-612
- PubMed: 30135578
- DOI: https://doi.org/10.1038/s41586-018-0440-4
- Primary Citation of Related Structures:
6GME, 6GMH - PubMed Abstract:
Gene regulation involves activation of RNA polymerase II (Pol II) that is paused and bound by the protein complexes DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF). Here we show that formation of an activated Pol II elongation complex in vitro requires the kinase function of the positive transcription elongation factor b (P-TEFb) and the elongation factors PAF1 complex (PAF) and SPT6. The cryo-EM structure of an activated elongation complex of Sus scrofa Pol II and Homo sapiens DSIF, PAF and SPT6 was determined at 3.1 Å resolution and compared to the structure of the paused elongation complex formed by Pol II, DSIF and NELF. PAF displaces NELF from the Pol II funnel for pause release. P-TEFb phosphorylates the Pol II linker to the C-terminal domain. SPT6 binds to the phosphorylated C-terminal-domain linker and opens the RNA clamp formed by DSIF. These results provide the molecular basis for Pol II pause release and elongation activation.
- Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Göttingen, Germany.
Organizational Affiliation: