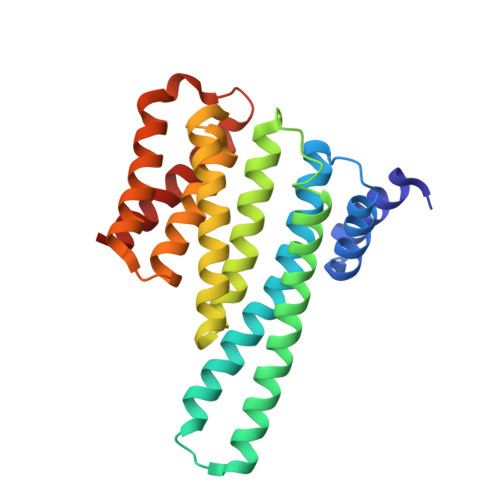
14-3-3 protein masks the nuclear localization sequence of caspase-2.
Smidova, A., Alblova, M., Kalabova, D., Psenakova, K., Rosulek, M., Herman, P., Obsil, T., Obsilova, V.(2018) FEBS J 285: 4196-4213
- PubMed: 30281929
- DOI: https://doi.org/10.1111/febs.14670
- Primary Citation of Related Structures:
6GKF, 6GKG - PubMed Abstract:
Caspase-2 is an apical protease responsible for the proteolysis of cellular substrates directly involved in mediating apoptotic signaling cascades. Caspase-2 activation is inhibited by phosphorylation followed by binding to the scaffolding protein 14-3-3, which recognizes two phosphoserines located in the linker between the caspase recruitment domain and the p19 domains of the caspase-2 zymogen. However, the structural details of this interaction and the exact role of 14-3-3 in the regulation of caspase-2 activation remain unclear. Moreover, the caspase-2 region with both 14-3-3-binding motifs also contains the nuclear localization sequence (NLS), thus suggesting that 14-3-3 binding may regulate the subcellular localization of caspase-2. Here, we report a structural analysis of the 14-3-3ζ:caspase-2 complex using a combined approach based on small angle X-ray scattering, NMR, chemical cross-linking, and fluorescence spectroscopy. The structural model proposed in this study suggests that phosphorylated caspase-2 and 14-3-3ζ form a compact and rigid complex in which the p19 and the p12 domains of caspase-2 are positioned within the central channel of the 14-3-3 dimer and stabilized through interactions with the C-terminal helices of both 14-3-3ζ protomers. In this conformation, the surface of the p12 domain, which is involved in caspase-2 activation by dimerization, is sterically occluded by the 14-3-3 dimer, thereby likely preventing caspase-2 activation. In addition, 14-3-3 protein binding to caspase-2 masks its NLS. Therefore, our results suggest that 14-3-3 protein binding to caspase-2 may play a key role in regulating caspase-2 activation. DATABASE: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.ww pdb.org (PDB ID codes 6GKF and 6GKG).
- Department of Structural Biology of Signaling Proteins, Division BIOCEV, Institute of Physiology of the Czech Academy of Sciences, Vestec, Czech Republic.
Organizational Affiliation: