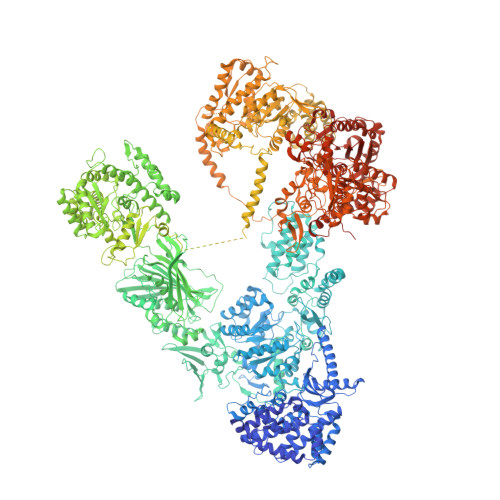
Structure of Type-I Mycobacterium tuberculosis fatty acid synthase at 3.3 angstrom resolution.
Elad, N., Baron, S., Peleg, Y., Albeck, S., Grunwald, J., Raviv, G., Shakked, Z., Zimhony, O., Diskin, R.(2018) Nat Commun 9: 3886-3886
- PubMed: 30250274
- DOI: https://doi.org/10.1038/s41467-018-06440-6
- Primary Citation of Related Structures:
6GJC - PubMed Abstract:
Tuberculosis (TB) is a devastating and rapidly spreading disease caused by Mycobacterium tuberculosis (Mtb). Therapy requires prolonged treatment with a combination of multiple agents and interruptions in the treatment regimen result in emergence and spread of multi-drug resistant (MDR) Mtb strains. MDR Mtb poses a significant global health problem, calling for urgent development of novel drugs to combat TB. Here, we report the 3.3 Å resolution structure of the ~2 MDa type-I fatty acid synthase (FAS-I) from Mtb, determined by single particle cryo-EM. Mtb FAS-I is an essential enzymatic complex that contributes to the virulence of Mtb, and thus a prime target for anti-TB drugs. The structural information for Mtb FAS-I we have obtained enables computer-based drug discovery approaches, and the resolution achieved by cryo-EM is sufficient for elucidating inhibition mechanisms by putative small molecular weight inhibitors.
- Department of Chemical Research Support, Weizmann Institute of Science, Rehovot, 7610001, Israel.
Organizational Affiliation: