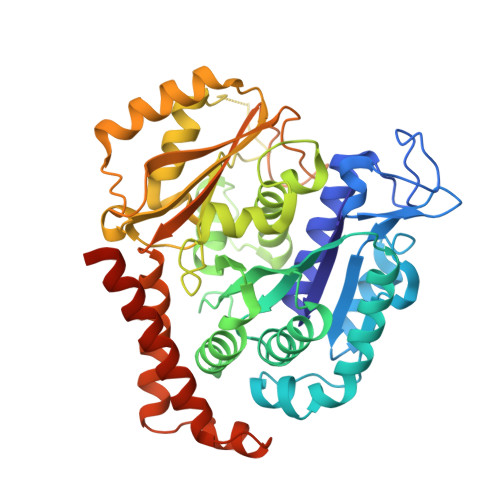
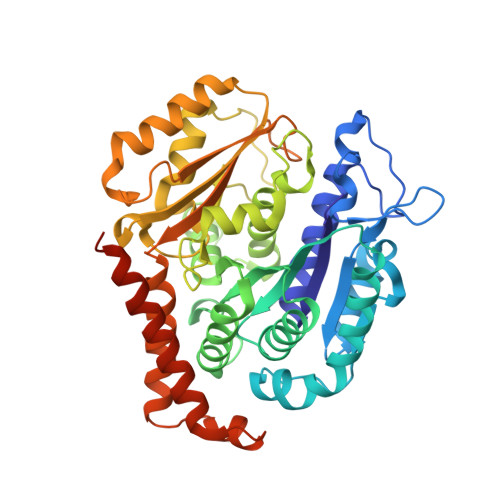
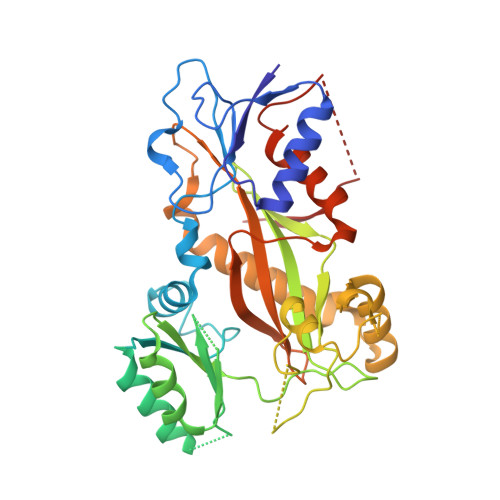
Structure-activity relationships, biological evaluation and structural studies of novel pyrrolonaphthoxazepines as antitumor agents.
Brindisi, M., Ulivieri, C., Alfano, G., Gemma, S., de Asis Balaguer, F., Khan, T., Grillo, A., Chemi, G., Menchon, G., Prota, A.E., Olieric, N., Lucena-Agell, D., Barasoain, I., Diaz, J.F., Nebbioso, A., Conte, M., Lopresti, L., Magnano, S., Amet, R., Kinsella, P., Zisterer, D.M., Ibrahim, O., O'Sullivan, J., Morbidelli, L., Spaccapelo, R., Baldari, C., Butini, S., Novellino, E., Campiani, G., Altucci, L., Steinmetz, M.O., Brogi, S.(2018) Eur J Med Chem 162: 290-320
- PubMed: 30448418
- DOI: https://doi.org/10.1016/j.ejmech.2018.11.004
- Primary Citation of Related Structures:
6GJ4 - PubMed Abstract:
Microtubule-targeting agents (MTAs) are a class of clinically successful anti-cancer drugs. The emergence of multidrug resistance to MTAs imposes the need for developing new MTAs endowed with diverse mechanistic properties. Benzoxazepines were recently identified as a novel class of MTAs. These anticancer agents were thoroughly characterized for their antitumor activity, although, their exact mechanism of action remained elusive. Combining chemical, biochemical, cellular, bioinformatics and structural efforts we developed improved pyrrolonaphthoxazepines antitumor agents and their mode of action at the molecular level was elucidated. Compound 6j, one of the most potent analogues, was confirmed by X-ray as a colchicine-site MTA. A comprehensive structural investigation was performed for a complete elucidation of the structure-activity relationships. Selected pyrrolonaphthoxazepines were evaluated for their effects on cell cycle, apoptosis and differentiation in a variety of cancer cells, including multidrug resistant cell lines. Our results define compound 6j as a potentially useful optimized hit for the development of effective compounds for treating drug-resistant tumors.
- European Research Centre for Drug Discovery and Development (NatSynDrugs), University of Siena, Department of Biotechnology, Chemistry and Pharmacy, DoE Department of Excellence 2018-2022, via Aldo Moro 2, I-53100, Siena, Italy; Istituto Toscano Tumori, University of Siena, via Aldo Moro 2, I-53100, Siena, Italy.
Organizational Affiliation: