Sustainable Syntheses of (-)-Jerantinines A & E and Structural Characterisation of the Jerantinine-Tubulin Complex at the Colchicine Binding Site.
Smedley, C.J., Stanley, P.A., Qazzaz, M.E., Prota, A.E., Olieric, N., Collins, H., Eastman, H., Barrow, A.S., Lim, K.H., Kam, T.S., Smith, B.J., Duivenvoorden, H.M., Parker, B.S., Bradshaw, T.D., Steinmetz, M.O., Moses, J.E.(2018) Sci Rep 8: 10617-10617
- PubMed: 30006510
- DOI: https://doi.org/10.1038/s41598-018-28880-2
- Primary Citation of Related Structures:
6GF3 - PubMed Abstract:
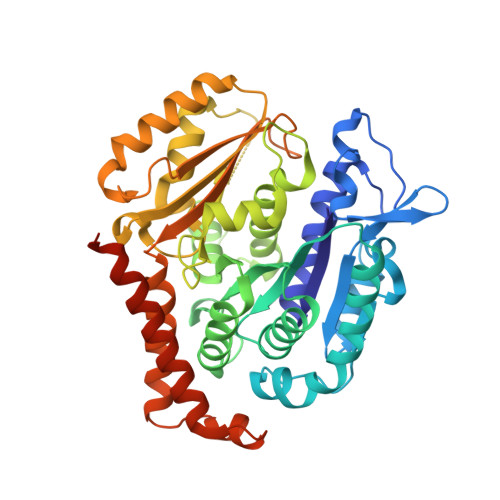
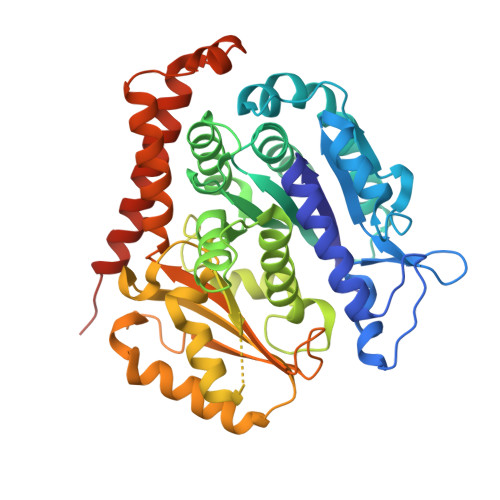
The jerantinine family of Aspidosperma indole alkaloids from Tabernaemontana corymbosa are potent microtubule-targeting agents with broad spectrum anticancer activity. The natural supply of these precious metabolites has been significantly disrupted due to the inclusion of T. corymbosa on the endangered list of threatened species by the International Union for Conservation of Nature. This report describes the asymmetric syntheses of (-)-jerantinines A and E from sustainably sourced (-)-tabersonine, using a straight-forward and robust biomimetic approach. Biological investigations of synthetic (-)-jerantinine A, along with molecular modelling and X-ray crystallography studies of the tubulin-(-)-jerantinine B acetate complex, advocate an anticancer mode of action of the jerantinines operating via microtubule disruption resulting from binding at the colchicine site. This work lays the foundation for accessing useful quantities of enantiomerically pure jerantinine alkaloids for future development.
- La Trobe Institute for Molecular Science, La Trobe University, Melbourne, VIC 3086, Australia.
Organizational Affiliation: